de.jstacs.sequenceScores.statisticalModels.trainable.hmm.models
Class HigherOrderHMM
java.lang.Object
 de.jstacs.sequenceScores.statisticalModels.trainable.AbstractTrainableStatisticalModel
de.jstacs.sequenceScores.statisticalModels.trainable.AbstractTrainableStatisticalModel
 de.jstacs.sequenceScores.statisticalModels.trainable.hmm.AbstractHMM
de.jstacs.sequenceScores.statisticalModels.trainable.hmm.AbstractHMM
 de.jstacs.sequenceScores.statisticalModels.trainable.hmm.models.HigherOrderHMM
de.jstacs.sequenceScores.statisticalModels.trainable.hmm.models.HigherOrderHMM
- All Implemented Interfaces:
- SequenceScore, StatisticalModel, TrainableStatisticalModel, Storable, Cloneable
- Direct Known Subclasses:
- DifferentiableHigherOrderHMM, SamplingHigherOrderHMM
public class HigherOrderHMM
- extends AbstractHMM
This class implements a higher order hidden Markov model.
Currently, the modeling of the transitions is higher order, but is easily possible to extend this to emissions.
This implementation allows to have a set of final states 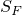 .
A state is denoted final states if it is allowed at the end of a path. Hence, any valid path always ends with a final state.
Using the method
.
A state is denoted final states if it is allowed at the end of a path. Hence, any valid path always ends with a final state.
Using the method AbstractTrainableStatisticalModel.getLogProbFor(Sequence) for sequence 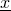 returns the value
returns the value
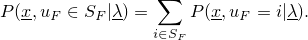
Setting 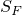 to all states leads to the computation of the likelihood.
to all states leads to the computation of the likelihood.
- Author:
- Jens Keilwagen
|
Nested Class Summary |
protected static class |
HigherOrderHMM.Type
This enum defined different types of computations that will be done using the backward algorithm. |
|
Field Summary |
protected double[] |
backwardIntermediate
Helper variable = only for internal use. |
protected int[] |
container
Helper variable = only for internal use. |
protected double[] |
logEmission
Helper variable = only for internal use. |
protected int[][] |
numberOfSummands
Helper variable = only for internal use. |
protected boolean |
skipInit
Indicates if the model should be initialized (randomly) before optimization |
protected IntList |
stateList
Helper variable = only for internal use. |
| Fields inherited from class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.AbstractHMM |
bwdMatrix, emission, emissionIdx, finalState, forward, fwdMatrix, name, sostream, START_NODE, states, threads, trainingParameter, transition |
|
Method Summary |
protected void |
appendFurtherInformation(StringBuffer xml)
This method appends further information to the XML representation. |
protected double |
baumWelch(int startPos,
int endPos,
double weight,
Sequence seq)
This method computes the likelihood and modifies the sufficient statistics according to the Baum-Welch algorithm. |
HigherOrderHMM |
clone()
Follows the conventions of Object's clone()-method. |
protected void |
createHelperVariables()
This method instantiates all helper variables that are need inside the model for instance for filling forward and backward matrix, ... |
protected void |
createStates()
This method creates states for the internal usage. |
protected void |
estimateFromStatistics()
This method estimates the parameters of all emissions and the transition using their sufficient statistics. |
protected void |
extractFurtherInformation(StringBuffer xml)
This method extracts further information from the XML representation. |
protected void |
fillBwdMatrix(int startPos,
int endPos,
Sequence seq)
This method fills the backward-matrix for a given sequence. |
protected void |
fillBwdOrViterbiMatrix(HigherOrderHMM.Type t,
int startPos,
int endPos,
double weight,
Sequence seq)
This method computes the entries of the backward or the viterbi matrix. |
protected void |
fillFwdMatrix(int startPos,
int endPos,
Sequence seq)
This method fills the forward-matrix for a given sequence. |
protected void |
fillLogStatePosteriorMatrix(double[][] statePosterior,
int startPos,
int endPos,
Sequence seq,
boolean silentZero)
This method fills the log state posterior of Sequence seq in a given matrix. |
protected void |
finalize()
|
ResultSet |
getCharacteristics()
Returns some information characterizing or describing the current
instance. |
String |
getInstanceName()
Should return a short instance name such as iMM(0), BN(2), ... |
double |
getLogPriorTerm()
Returns a value that is proportional to the log of the prior. |
double |
getLogProbForPath(IntList path,
int startPos,
Sequence seq)
|
double[] |
getLogScoreFor(DataSet data)
This method computes the logarithm of the scores of all sequences
in the given sample. |
void |
getLogScoreFor(DataSet data,
double[] res)
This method computes and stores the logarithm of the scores for
any sequence in the sample in the given double-array. |
byte |
getMaximalMarkovOrder()
This method returns the maximal used Markov order, if possible. |
NumericalResultSet |
getNumericalCharacteristics()
Returns the subset of numerical values that are also returned by
SequenceScore.getCharacteristics(). |
Pair<IntList,Double> |
getViterbiPathFor(int startPos,
int endPos,
Sequence seq)
|
protected String |
getXMLTag()
Returns the tag for the XML representation. |
protected void |
initialize(DataSet data,
double[] weight)
This method initializes all emissions and the transition. |
protected void |
initializeRandomly()
This method initializes all emissions and the transition randomly. |
boolean |
isInitialized()
This method can be used to determine whether the instance is initialized. |
protected void |
resetStatistics()
This method resets all sufficient statistics of all emissions and the transition. |
void |
samplePath(IntList path,
int startPos,
int endPos,
Sequence seq)
This method samples a valid path for the given sequence seq using the internal parameters. |
void |
train(DataSet data,
double[] weights)
Trains the TrainableStatisticalModel object given the data as DataSet using
the specified weights. |
protected double |
viterbi(IntList path,
int startPos,
int endPos,
double weight,
Sequence seq)
This method computes the viterbi score of a given sequence seq. |
| Methods inherited from class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.AbstractHMM |
createMatrixForStatePosterior, decodePath, decodeStatePosterior, determineFinalStates, fromXML, getFinalStatePosterioriMatrix, getGraphvizRepresentation, getGraphvizRepresentation, getGraphvizRepresentation, getGraphvizRepresentation, getLogProbFor, getLogStatePosteriorMatrixFor, getLogStatePosteriorMatrixFor, getNumberOfStates, getNumberOfThreads, getRunTimeException, getStatePosteriorMatrixFor, getStatePosteriorMatrixFor, getViterbiPathFor, getViterbiPathsFor, initTransition, logProb, provideMatrix, setOutputStream, toString, toXML, train |
container
protected int[] container
- Helper variable = only for internal use. This array is used for
Transition.fillTransitionInformation(int, int, int, int[]).
logEmission
protected double[] logEmission
- Helper variable = only for internal use. This array is used for compute the emissions at each position of a sequence only once,
which might be beneficial for higher order models.
- See Also:
AbstractHMM.emission
backwardIntermediate
protected double[] backwardIntermediate
- Helper variable = only for internal use. This array is used to compute the backward matrix. It stores intermediate results.
#see
numberOfSummands
numberOfSummands
protected int[][] numberOfSummands
- Helper variable = only for internal use. This array is used to compute the forward and backward matrix. It stores the number of intermediate results.
stateList
protected IntList stateList
- Helper variable = only for internal use. This field is used in the method
samplePath(IntList, int, int, Sequence).
skipInit
protected boolean skipInit
- Indicates if the model should be initialized (randomly) before optimization
HigherOrderHMM
public HigherOrderHMM(HMMTrainingParameterSet trainingParameterSet,
String[] name,
Emission[] emission,
BasicHigherOrderTransition.AbstractTransitionElement... te)
throws Exception
- This is a convenience constructor. It assumes that state
i used emission i on the forward strand.
- Parameters:
trainingParameterSet - the ParameterSet that determines the training algorithm and contains the necessary Parametersname - the names of the statesemission - the emissionste - the BasicHigherOrderTransition.AbstractTransitionElements building a transition
- Throws:
Exception - if
- some component could not be cloned
- some the length of
name, emissionIdx, or forward is not equal to the number of states
- not all emissions use the same
AlphabetContainer
- the states can not be handled by the transition
- See Also:
HigherOrderHMM(HMMTrainingParameterSet, String[], int[], boolean[], Emission[], de.jstacs.sequenceScores.statisticalModels.trainable.hmm.transitions.BasicHigherOrderTransition.AbstractTransitionElement...)
HigherOrderHMM
public HigherOrderHMM(HMMTrainingParameterSet trainingParameterSet,
String[] name,
int[] emissionIdx,
boolean[] forward,
Emission[] emission,
BasicHigherOrderTransition.AbstractTransitionElement... te)
throws Exception
- This is the main constructor.
- Parameters:
trainingParameterSet - the ParameterSet that determines the training algorithm and contains the necessary Parametersname - the names of the statesemissionIdx - the indices of the emissions that should be used for each state, if null state i will use emission iforward - a boolean array that indicates whether the symbol on the forward or the reverse complementary strand should be used,
if null all states use the forward strandemission - the emissionste - the BasicHigherOrderTransition.AbstractTransitionElements building a transition
- Throws:
Exception - if
- some component could not be cloned
- some the length of
name, emissionIdx, or forward is not equal to the number of states
- not all emissions use the same
AlphabetContainer
- the states can not be handled by the transition
HigherOrderHMM
public HigherOrderHMM(StringBuffer xml)
throws NonParsableException
- The standard constructor for the interface
Storable.
Constructs an HigherOrderHMM out of an XML representation.
- Parameters:
xml - the XML representation as StringBuffer
- Throws:
NonParsableException - if the HigherOrderHMM could not be reconstructed out of
the StringBuffer xml
createHelperVariables
protected void createHelperVariables()
- Description copied from class:
AbstractHMM
- This method instantiates all helper variables that are need inside the model for instance for filling forward and backward matrix, ...
- Specified by:
createHelperVariables in class AbstractHMM
getXMLTag
protected String getXMLTag()
- Description copied from class:
AbstractHMM
- Returns the tag for the XML representation.
- Specified by:
getXMLTag in class AbstractHMM
- Returns:
- the tag for the XML representation
- See Also:
AbstractHMM.fromXML(StringBuffer),
AbstractHMM.toXML()
appendFurtherInformation
protected void appendFurtherInformation(StringBuffer xml)
- Description copied from class:
AbstractHMM
- This method appends further information to the XML representation. It allows subclasses to save further parameters that are not defined in the superclass.
- Specified by:
appendFurtherInformation in class AbstractHMM
- Parameters:
xml - the XML representation
extractFurtherInformation
protected void extractFurtherInformation(StringBuffer xml)
throws NonParsableException
- This method extracts further information from the XML representation. It allows subclasses to cast further parameters that are not defined in the superclass.
- Specified by:
extractFurtherInformation in class AbstractHMM
- Parameters:
xml - the XML representation
- Throws:
NonParsableException - if the information could not be reconstructed out of the StringBuffer xml
clone
public HigherOrderHMM clone()
throws CloneNotSupportedException
- Description copied from class:
AbstractTrainableStatisticalModel
- Follows the conventions of
Object's clone()-method.
- Specified by:
clone in interface SequenceScore- Specified by:
clone in interface TrainableStatisticalModel- Overrides:
clone in class AbstractHMM
- Returns:
- an object, that is a copy of the current
AbstractTrainableStatisticalModel
(the member-AlphabetContainer isn't deeply cloned since
it is assumed to be immutable). The type of the returned object
is defined by the class X directly inherited from
AbstractTrainableStatisticalModel. Hence X's
clone()-method should work as:
1. Object o = (X)super.clone();
2. all additional member variables of o defined by
X that are not of simple data-types like
int, double, ... have to be deeply
copied
3. return o
- Throws:
CloneNotSupportedException - if something went wrong while cloning
createStates
protected void createStates()
- Description copied from class:
AbstractHMM
- This method creates states for the internal usage.
- Specified by:
createStates in class AbstractHMM
getLogPriorTerm
public double getLogPriorTerm()
- Description copied from interface:
StatisticalModel
- Returns a value that is proportional to the log of the prior. For maximum likelihood (ML) 0
should be returned.
- Returns:
- a value that is proportional to the log of the prior
getLogProbForPath
public double getLogProbForPath(IntList path,
int startPos,
Sequence seq)
throws Exception
- Specified by:
getLogProbForPath in class AbstractHMM
- Parameters:
path - the given state pathstartPos - the start position within the sequence(s) (inclusive)seq - the sequence(s)
- Returns:
- the logarithm of the probability for the given path and the given sequence(s)
- Throws:
Exception - if the probability for the sequence given path could not be computed, for instance if the model is not trained, ...
fillLogStatePosteriorMatrix
protected void fillLogStatePosteriorMatrix(double[][] statePosterior,
int startPos,
int endPos,
Sequence seq,
boolean silentZero)
throws Exception
- Description copied from class:
AbstractHMM
- This method fills the log state posterior of Sequence
seq in a given matrix.
- Specified by:
fillLogStatePosteriorMatrix in class AbstractHMM
- Parameters:
statePosterior - the matrix for the log state posteriorstartPos - the start positionendPos - the end positionseq - the sequencesilentZero - true if the state posterior for silent states is defined to be zero, otherwise false
- Throws:
Exception - if an error occurs during the computation- See Also:
AbstractHMM.getLogStatePosteriorMatrixFor(int, int, Sequence),
AbstractHMM.createMatrixForStatePosterior(int, int)
fillFwdMatrix
protected void fillFwdMatrix(int startPos,
int endPos,
Sequence seq)
throws OperationNotSupportedException,
WrongLengthException
- Description copied from class:
AbstractHMM
- This method fills the forward-matrix for a given sequence.
- Specified by:
fillFwdMatrix in class AbstractHMM
- Parameters:
startPos - the start position (inclusive) in the sequenceendPos - the end position (inclusive) in the sequenceseq - the sequence
- Throws:
OperationNotSupportedException
WrongLengthException
fillBwdMatrix
protected void fillBwdMatrix(int startPos,
int endPos,
Sequence seq)
throws Exception
- Description copied from class:
AbstractHMM
- This method fills the backward-matrix for a given sequence.
- Specified by:
fillBwdMatrix in class AbstractHMM
- Parameters:
startPos - the start position (inclusive) in the sequenceendPos - the end position (inclusive) in the sequenceseq - the sequence
- Throws:
Exception - if some error occurs during the computation
fillBwdOrViterbiMatrix
protected void fillBwdOrViterbiMatrix(HigherOrderHMM.Type t,
int startPos,
int endPos,
double weight,
Sequence seq)
throws Exception
- This method computes the entries of the backward or the viterbi matrix.
Additionally it allows to modify the sufficient statistics as needed for Baum-Welch training.
- Parameters:
t - a switch to decide which computation modestartPos - start position of the sequenceendPos - end position of the sequenceweight - the given external weight of the sequence (only used for Baum-Welch)seq - the sequence
- Throws:
Exception - forwarded from TrainableState.addToStatistic(int, int, double, de.jstacs.data.sequences.Sequence) and State.getLogScoreFor(int, int, Sequence)
getViterbiPathFor
public Pair<IntList,Double> getViterbiPathFor(int startPos,
int endPos,
Sequence seq)
throws Exception
- Specified by:
getViterbiPathFor in class AbstractHMM
- Parameters:
startPos - the start position within the sequenceendPos - the end position within the sequenceseq - the sequence
- Returns:
- a
Pair containing the viterbi state path and the corresponding score
- Throws:
Exception - if the viterbi path could not be computed, for instance if the model is not trained, ...
viterbi
protected double viterbi(IntList path,
int startPos,
int endPos,
double weight,
Sequence seq)
throws Exception
- This method computes the viterbi score of a given sequence
seq.
Furthermore, it allows either to modify the sufficient statistics according
to the viterbi training algorithm or to compute the viterbi path, which will
in this case be returned in path.
- Parameters:
path - if null viterbi training, otherwise computation of the viterbi pathstartPos - the start positionendPos - the end positionweight - the sequence weight, in most cases this is 1seq - the sequence
- Returns:
- the viterbi score of the sequence
- Throws:
Exception - an error occurs during the computation
baumWelch
protected double baumWelch(int startPos,
int endPos,
double weight,
Sequence seq)
throws Exception
- This method computes the likelihood and modifies the sufficient statistics according to the Baum-Welch algorithm.
- Parameters:
startPos - the start positionendPos - the end positionweight - the sequence weight, in most cases this is 1seq - the sequence
- Returns:
- the likelihood of the sequence
- Throws:
Exception - an error occurs during the computation
train
public void train(DataSet data,
double[] weights)
throws Exception
- Description copied from interface:
TrainableStatisticalModel
- Trains the
TrainableStatisticalModel object given the data as DataSet using
the specified weights. The weight at position i belongs to the element at
position i. So the array weight should have the number of
sequences in the sample as dimension. (Optionally it is possible to use
weight == null if all weights have the value one.)
This method should work non-incrementally. That means the result of the
following series: train(data1); train(data2)
should be a fully trained model over data2 and not over
data1+data2. All parameters of the model were given by the
call of the constructor.
- Parameters:
data - the given sequences as DataSetweights - the weights of the elements, each weight should be
non-negative
- Throws:
Exception - if the training did not succeed (e.g. the dimension of
weights and the number of sequences in the
sample do not match)- See Also:
DataSet.getElementAt(int),
DataSet.ElementEnumerator
initialize
protected void initialize(DataSet data,
double[] weight)
throws Exception
- This method initializes all emissions and the transition.
The initialization might use the data, but the default
implementation refers to
initializeRandomly().
- Parameters:
data - the data setweight - the weights for each sequence of the data set
- Throws:
Exception - if an error occurs during the initialization
initializeRandomly
protected void initializeRandomly()
- This method initializes all emissions and the transition randomly.
resetStatistics
protected void resetStatistics()
- This method resets all sufficient statistics of all emissions and the transition.
estimateFromStatistics
protected void estimateFromStatistics()
- This method estimates the parameters of all emissions and the transition using their sufficient statistics.
getMaximalMarkovOrder
public final byte getMaximalMarkovOrder()
throws UnsupportedOperationException
- Description copied from interface:
StatisticalModel
- This method returns the maximal used Markov order, if possible.
- Specified by:
getMaximalMarkovOrder in interface StatisticalModel- Overrides:
getMaximalMarkovOrder in class AbstractTrainableStatisticalModel
- Returns:
- maximal used Markov order
- Throws:
UnsupportedOperationException - if the model can't give a proper answer
getCharacteristics
public ResultSet getCharacteristics()
throws Exception
- Description copied from interface:
SequenceScore
- Returns some information characterizing or describing the current
instance. This could be e.g. the number of edges for a
Bayesian network or an image showing some representation of the instance.
The set of characteristics should always include the XML-representation
of the instance. The corresponding result type is
StorableResult.
- Specified by:
getCharacteristics in interface SequenceScore- Overrides:
getCharacteristics in class AbstractTrainableStatisticalModel
- Returns:
- the characteristics of the current instance
- Throws:
Exception - if some of the characteristics could not be defined- See Also:
StorableResult
getInstanceName
public String getInstanceName()
- Description copied from interface:
SequenceScore
- Should return a short instance name such as iMM(0), BN(2), ...
- Returns:
- a short instance name
getLogScoreFor
public double[] getLogScoreFor(DataSet data)
throws Exception
- Description copied from interface:
SequenceScore
- This method computes the logarithm of the scores of all sequences
in the given sample. The values are stored in an array according to the
index of the respective sequence in the sample.
The score for any sequence shall be computed independent of all
other sequences in the sample. So the result should be exactly the same
as for the method SequenceScore.getLogScoreFor(Sequence).
- Specified by:
getLogScoreFor in interface SequenceScore- Overrides:
getLogScoreFor in class AbstractTrainableStatisticalModel
- Parameters:
data - the sample of sequences
- Returns:
- an array containing the logarithm of the score of all
sequences of the sample
- Throws:
Exception - if something went wrong- See Also:
SequenceScore.getLogScoreFor(Sequence)
getLogScoreFor
public void getLogScoreFor(DataSet data,
double[] res)
throws Exception
- Description copied from interface:
SequenceScore
- This method computes and stores the logarithm of the scores for
any sequence in the sample in the given
double-array.
The score for any sequence shall be computed independent of all
other sequences in the sample. So the result should be exactly the same
as for the method SequenceScore.getLogScoreFor(Sequence).
- Specified by:
getLogScoreFor in interface SequenceScore- Overrides:
getLogScoreFor in class AbstractTrainableStatisticalModel
- Parameters:
data - the sample of sequencesres - the array for the results, has to have length
data.getNumberOfElements() (which returns the
number of sequences in the sample)
- Throws:
Exception - if something went wrong- See Also:
SequenceScore.getLogScoreFor(Sequence),
SequenceScore.getLogScoreFor(DataSet)
getNumericalCharacteristics
public NumericalResultSet getNumericalCharacteristics()
throws Exception
- Description copied from interface:
SequenceScore
- Returns the subset of numerical values that are also returned by
SequenceScore.getCharacteristics().
- Returns:
- the numerical characteristics of the current instance
- Throws:
Exception - if some of the characteristics could not be defined
isInitialized
public boolean isInitialized()
- Description copied from interface:
SequenceScore
- This method can be used to determine whether the instance is initialized. If
the instance is initialized you should be able to invoke
SequenceScore.getLogScoreFor(Sequence).
- Returns:
true if the instance is initialized, false
otherwise
finalize
protected void finalize()
throws Throwable
- Overrides:
finalize in class AbstractHMM
- Throws:
Throwable
samplePath
public void samplePath(IntList path,
int startPos,
int endPos,
Sequence seq)
throws Exception
- This method samples a valid path for the given sequence
seq using the internal parameters.
- Parameters:
path - an IntList containing the path after using this methodstartPos - the start positionendPos - the end positionseq - the sequence
- Throws:
Exception - if an error occurs during computation
 de.jstacs.sequenceScores.statisticalModels.trainable.AbstractTrainableStatisticalModel
de.jstacs.sequenceScores.statisticalModels.trainable.AbstractTrainableStatisticalModel
 de.jstacs.sequenceScores.statisticalModels.trainable.hmm.AbstractHMM
de.jstacs.sequenceScores.statisticalModels.trainable.hmm.AbstractHMM
 de.jstacs.sequenceScores.statisticalModels.trainable.hmm.models.HigherOrderHMM
de.jstacs.sequenceScores.statisticalModels.trainable.hmm.models.HigherOrderHMM
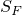 .
A state is denoted final states if it is allowed at the end of a path. Hence, any valid path always ends with a final state.
Using the method
.
A state is denoted final states if it is allowed at the end of a path. Hence, any valid path always ends with a final state.
Using the method 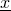 returns the value
returns the value