- L - Variable in class de.jstacs.algorithms.graphs.tensor.Tensor
-
The number of nodes minus 1.
- l1 - Variable in class de.jstacs.algorithms.alignment.Alignment
-
The length of the sub-sequence of the first sequence that is aligned
- l2 - Variable in class de.jstacs.algorithms.alignment.Alignment
-
The length of the sub-sequence of the second sequence that is aligned
- LargeSequenceReader - Class in de.jstacs.utils
-
Class for reading large DNA sequences (e.g.
- LargeSequenceReader() - Constructor for class de.jstacs.utils.LargeSequenceReader
-
- lastParameters - Variable in class de.jstacs.classifiers.differentiableSequenceScoreBased.sampling.SamplingScoreBasedClassifier
-
The last accepted parameters for all samplings, backup for iterative
sampling when checking for
BurnInTest
- lastScore - Variable in class de.jstacs.classifiers.differentiableSequenceScoreBased.sampling.SamplingScoreBasedClassifier
-
The scores yielded for the parameters in lastParameters
- leafOrder(double[][]) - Method in class de.jstacs.clustering.hierachical.ClusterTree
-
Orders the leaves of this cluster tree such that adjacent nodes have minimal distance.
- LearningPrinciple - Enum in de.jstacs.classifiers.differentiableSequenceScoreBased.gendismix
-
This enum can be used to obtain the weights for well-known optimization tasks.
- length() - Method in class de.jstacs.data.alphabets.Alphabet
-
- length() - Method in class de.jstacs.data.alphabets.ContinuousAlphabet
-
- length() - Method in class de.jstacs.data.alphabets.DiscreteAlphabet
-
Returns the number of symbols in the calling alphabet.
- length - Variable in class de.jstacs.parameters.SequenceScoringParameterSet
-
The length of sequences the model can work on or 0 for
arbitrary length
- length - Variable in class de.jstacs.sequenceScores.differentiable.AbstractDifferentiableSequenceScore
-
The length of the modeled sequences.
- length - Variable in class de.jstacs.sequenceScores.statisticalModels.trainable.AbstractTrainableStatisticalModel
-
The length of the sequences the model can classify.
- length() - Method in class de.jstacs.utils.DoubleList
-
Returns the number of inserted elements.
- length() - Method in class de.jstacs.utils.IntList
-
Returns the number of inserted elements.
- lengths - Variable in class de.jstacs.sequenceScores.statisticalModels.trainable.CompositeTrainSM
-
The length for each component.
- LEQ - Static variable in interface de.jstacs.parameters.validation.Constraint
-
The condition is less or equal
- LIKELIHOOD_INDEX - Static variable in enum de.jstacs.classifiers.differentiableSequenceScoreBased.gendismix.LearningPrinciple
-
This constant is the array index of the weighting factor for the likelihood.
- LimitedMedianStartDistance - Class in de.jstacs.algorithms.optimization
-
- LimitedMedianStartDistance(int, double) - Constructor for class de.jstacs.algorithms.optimization.LimitedMedianStartDistance
-
This constructor creates an instance with slots memory slots
that will initially be filled with value.
- limitedMemoryBFGS(DifferentiableFunction, double[], byte, TerminationCondition, double, StartDistanceForecaster, OutputStream, Time) - Static method in class de.jstacs.algorithms.optimization.Optimizer
-
The Broyden-Fletcher-Goldfarb-Shanno version
of limited memory quasi-Newton methods.
- LimitedSparseLocalInhomogeneousMixtureDiffSM_higherOrder - Class in de.jstacs.sequenceScores.statisticalModels.differentiable.localMixture
-
Class for a sparse local inhomogeneous mixture (Slim) model.
- LimitedSparseLocalInhomogeneousMixtureDiffSM_higherOrder(AlphabetContainer, int, int, int, double, double, LimitedSparseLocalInhomogeneousMixtureDiffSM_higherOrder.PriorType) - Constructor for class de.jstacs.sequenceScores.statisticalModels.differentiable.localMixture.LimitedSparseLocalInhomogeneousMixtureDiffSM_higherOrder
-
Creates a new Slim model with given number of components and maximum distance.
- LimitedSparseLocalInhomogeneousMixtureDiffSM_higherOrder(StringBuffer) - Constructor for class de.jstacs.sequenceScores.statisticalModels.differentiable.localMixture.LimitedSparseLocalInhomogeneousMixtureDiffSM_higherOrder
-
- LimitedSparseLocalInhomogeneousMixtureDiffSM_higherOrder.PriorType - Enum in de.jstacs.sequenceScores.statisticalModels.differentiable.localMixture
-
The type of the prior used by the Slim model
- LimitedStringExtractor - Class in de.jstacs.io
-
- LimitedStringExtractor(AbstractStringExtractor, int) - Constructor for class de.jstacs.io.LimitedStringExtractor
-
This constructor creates a new instance based on a given
AbstractStringExtractor and a maximal number of Strings to be read.
- LIN_EPS - Static variable in class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.inhomogeneous.MEMTools
-
The epsilon for the line search in an optimization using the
Optimizer.
- linearizeParameters() - Method in class de.jstacs.sequenceScores.statisticalModels.differentiable.directedGraphicalModels.BNDiffSMParameterTree
-
Extracts the
BNDiffSMParameters from the leaves of this tree in
left-to-right order (as specified by the order of the alphabet) and
returns them as a
LinkedList.
- LineBasedResult(String, String, DataType) - Constructor for class de.jstacs.tools.ui.galaxy.GalaxyAdaptor.LineBasedResult
-
- LinkedImageResult(String, String, BufferedImage, GalaxyAdaptor.FileResult) - Constructor for class de.jstacs.tools.ui.galaxy.GalaxyAdaptor.LinkedImageResult
-
- LinkedImageResult(StringBuffer) - Constructor for class de.jstacs.tools.ui.galaxy.GalaxyAdaptor.LinkedImageResult
-
- list - Variable in class de.jstacs.results.ListResult
-
- ListResult - Class in de.jstacs.results
-
Class for a
Result that contains a list or a matrix, respectively, of
ResultSets.
- ListResult(String, String, ResultSet, ResultSet...) - Constructor for class de.jstacs.results.ListResult
-
- ListResult(String, String, ResultSet, Collection<ResultSet>) - Constructor for class de.jstacs.results.ListResult
-
- ListResult(StringBuffer) - Constructor for class de.jstacs.results.ListResult
-
The standard constructor for the interface
Storable.
- ListResultSaver - Class in de.jstacs.results.savers
-
- llGrad - Variable in class de.jstacs.classifiers.differentiableSequenceScoreBased.gendismix.LogGenDisMixFunction
-
Array for the gradient of the log-likelihood
- LocatedSequenceAnnotation - Class in de.jstacs.data.sequences.annotation
-
Class for a
SequenceAnnotation that has a position on the sequence,
e.g for transcription start sites or intron-exon junctions.
- LocatedSequenceAnnotation(int, String, String, Result...) - Constructor for class de.jstacs.data.sequences.annotation.LocatedSequenceAnnotation
-
- LocatedSequenceAnnotation(int, String, String, Collection<Result>) - Constructor for class de.jstacs.data.sequences.annotation.LocatedSequenceAnnotation
-
- LocatedSequenceAnnotation(int, String, String, SequenceAnnotation[], Result...) - Constructor for class de.jstacs.data.sequences.annotation.LocatedSequenceAnnotation
-
- LocatedSequenceAnnotation(String, String, LocatedSequenceAnnotation[], Result...) - Constructor for class de.jstacs.data.sequences.annotation.LocatedSequenceAnnotation
-
- LocatedSequenceAnnotation(StringBuffer) - Constructor for class de.jstacs.data.sequences.annotation.LocatedSequenceAnnotation
-
The standard constructor for the interface
Storable.
- LocatedSequenceAnnotationWithLength - Class in de.jstacs.data.sequences.annotation
-
- LocatedSequenceAnnotationWithLength(int, int, String, String, Result...) - Constructor for class de.jstacs.data.sequences.annotation.LocatedSequenceAnnotationWithLength
-
- LocatedSequenceAnnotationWithLength(int, int, String, String, Collection<Result>) - Constructor for class de.jstacs.data.sequences.annotation.LocatedSequenceAnnotationWithLength
-
- LocatedSequenceAnnotationWithLength(int, int, String, String, SequenceAnnotation[], Result...) - Constructor for class de.jstacs.data.sequences.annotation.LocatedSequenceAnnotationWithLength
-
- LocatedSequenceAnnotationWithLength(String, String, LocatedSequenceAnnotation[], Result...) - Constructor for class de.jstacs.data.sequences.annotation.LocatedSequenceAnnotationWithLength
-
- LocatedSequenceAnnotationWithLength(StringBuffer) - Constructor for class de.jstacs.data.sequences.annotation.LocatedSequenceAnnotationWithLength
-
The standard constructor for the interface
Storable.
- logClazz - Variable in class de.jstacs.classifiers.differentiableSequenceScoreBased.AbstractOptimizableFunction
-
The logarithm of the class parameters.
- logEmission - Variable in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.models.HigherOrderHMM
-
Helper variable = only for internal use.
- logGammaSum - Variable in class de.jstacs.sequenceScores.statisticalModels.differentiable.mixture.AbstractMixtureDiffSM
-
This double contains the sum of the logarithm of the gamma
functions used in the prior.
- LogGenDisMixFunction - Class in de.jstacs.classifiers.differentiableSequenceScoreBased.gendismix
-
This class implements the the following function
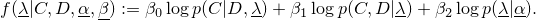
The weights
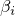
have to sum to 1.
- LogGenDisMixFunction(int, DifferentiableSequenceScore[], DataSet[], double[][], LogPrior, double[], boolean, boolean) - Constructor for class de.jstacs.classifiers.differentiableSequenceScoreBased.gendismix.LogGenDisMixFunction
-
The constructor for creating an instance that can be used in an
Optimizer.
- logHiddenNorm - Variable in class de.jstacs.sequenceScores.statisticalModels.differentiable.mixture.AbstractMixtureDiffSM
-
This double contains the logarithm of the normalization
constant of hidden parameters of the instance.
- logHiddenPotential - Variable in class de.jstacs.sequenceScores.statisticalModels.differentiable.mixture.AbstractMixtureDiffSM
-
This array contains the logarithm of the hidden potentials of the
instance.
- LogisticConstraint - Interface in de.jstacs.sequenceScores.differentiable.logistic
-
This interface defines the function
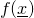
for sequence
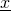
which can be used in
LogisticDiffSS.
- LogisticDiffSS - Class in de.jstacs.sequenceScores.differentiable.logistic
-
This class implements a logistic function.
- LogisticDiffSS(AlphabetContainer, int, LogisticConstraint...) - Constructor for class de.jstacs.sequenceScores.differentiable.logistic.LogisticDiffSS
-
- LogisticDiffSS(StringBuffer) - Constructor for class de.jstacs.sequenceScores.differentiable.logistic.LogisticDiffSS
-
- logNorm - Variable in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.states.emissions.discrete.AbstractConditionalDiscreteEmission
-
The log-normalization constants for each condition
- logNorm - Variable in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.transitions.BasicHigherOrderTransition.AbstractTransitionElement
-
The log normalization constant based on the parameters.
- logNormalizationConstant - Variable in class de.jstacs.sequenceScores.statisticalModels.differentiable.directedGraphicalModels.BayesianNetworkDiffSM
-
Normalization constant to obtain normalized probabilities.
- LogPrior - Class in de.jstacs.classifiers.differentiableSequenceScoreBased.logPrior
-
The abstract class for any log-prior used e.g.
- LogPrior() - Constructor for class de.jstacs.classifiers.differentiableSequenceScoreBased.logPrior.LogPrior
-
- logProb(int, int, Sequence) - Method in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.AbstractHMM
-
This method computes the logarithm of the probability of the corresponding subsequences.
- logProb(int, int, Sequence) - Method in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.models.DifferentiableHigherOrderHMM
-
- logProb(int, int, Sequence) - Method in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.models.SamplingHigherOrderHMM
-
- logProbFor(Sequence, int, int) - Method in class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.homogeneous.HomogeneousMM
-
- logProbFor(Sequence, int, int) - Method in class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.homogeneous.HomogeneousTrainSM
-
This method computes the logarithm of the probability of the given
Sequence in the given interval.
- logSumNormalisation(double[]) - Static method in class de.jstacs.utils.Normalisation
-
The method does a log-sum-normalisation on the array d, where
the values of d are assumed to be logarithmised.
- logSumNormalisation(double[], int, int) - Static method in class de.jstacs.utils.Normalisation
-
The method does a log-sum-normalisation on the values of the array d between start
index startD and end index endD, where
the values of d are assumed to be logarithmised.
- logSumNormalisation(double[], int, int, double[]) - Static method in class de.jstacs.utils.Normalisation
-
The method does a log-sum-normalisation on the values of the array d between start
index startD and end index endD, where
the values of d are assumed to be logarithmised.
- logSumNormalisation(double[], int, int, double[], int) - Static method in class de.jstacs.utils.Normalisation
-
The method does a log-sum-normalisation on the values of the array d between start
index startD and end index endD, where
the values of d are assumed to be logarithmised.
- logSumNormalisation(double[], int, int, double[], double[], int) - Static method in class de.jstacs.utils.Normalisation
-
The method does a log-sum-normalisation on the values of the array d between start
index startD and end index endD, where
the values of d are assumed to be logarithmised.
- logSumNormalisation(double[], double) - Static method in class de.jstacs.utils.Normalisation
-
The method does a log-sum-normalisation on the array d, where
the values of d are assumed to be logarithmised.
- logSumNormalisation(double[], int, int, double, double[], int) - Static method in class de.jstacs.utils.Normalisation
-
The method does a log-sum-normalisation on the values of the array d between start
index startD and end index endD, where
the values of d are assumed to be logarithmised.
- logSumNormalisation(double[], int, int, double, double[], double[], int) - Static method in class de.jstacs.utils.Normalisation
-
The method does a log-sum-normalisation on the values of the array d between start
index startD and end index endD, where
the values of d are assumed to be logarithmised.
- logWeights - Variable in class de.jstacs.sequenceScores.statisticalModels.trainable.mixture.AbstractMixtureTrainSM
-
The log probabilities for each component.
- lookup - Variable in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.transitions.BasicHigherOrderTransition
-
The lookup table for spare context en- and decoding.
- LT - Static variable in interface de.jstacs.parameters.validation.Constraint
-
The condition is less than
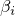 have to sum to 1.
have to sum to 1.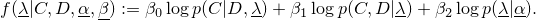
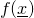 for sequence
for sequence 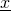 which can be used in
which can be used in