- java.lang.Object
-
- de.jstacs.sequenceScores.statisticalModels.trainable.hmm.states.emissions.SilentEmission
-
- All Implemented Interfaces:
- SamplingComponent, SamplingFromStatistic, DifferentiableEmission, Emission, SamplingEmission, Storable, Cloneable
public final class SilentEmission extends Object implements DifferentiableEmission, SamplingEmission
This class implements a silent emission which is used to create silent states.- Author:
- Jens Keilwagen
-
-
Constructor Summary
Constructors Constructor and Description SilentEmission()The main constructor.SilentEmission(StringBuffer xml)The standard constructor for the interfaceStorable.
-
Method Summary
All Methods Instance Methods Concrete Methods Modifier and Type Method and Description voidacceptParameters()This methods accepts the drawn parameters.voidaddGradientOfLogPriorTerm(double[] gradient, int offset)This method computes the gradient ofEmission.getLogPriorTerm()for each parameter of this model.voidaddToStatistic(boolean forward, int startPos, int endPos, double weight, Sequence seq)This method adds theweightto the internal sufficient statistic.SilentEmissionclone()voiddrawParametersFromStatistic()This method draws the parameters using a sufficient statistic representing a posteriori density.voidestimateFromStatistic()This method estimates the parameters from the internal sufficient statistic.voidextendSampling(int sampling, boolean append)This method allows to extend a sampling.voidfillCurrentParameter(double[] params)Fills the current parameters in the globalparamsarray using the internal offset.voidfillSamplingGroups(int parameterOffset, LinkedList<int[]> list)Adds the groups of indexes of those parameters of this emission that should be sampled together in one step of a grouped sampling procedure, each as anint[], intolist.AlphabetContainergetAlphabetContainer()This method returns theAlphabetContainerof this emission.doublegetLogGammaScoreFromStatistic()This method calculates a score for the current statistics, which is independent from the current parameters In general the gamma-score is a product of gamma-functions parameterized with the current statisticsdoublegetLogPosteriorFromStatistic()This method calculates the a-posteriori probability for the current statisticsdoublegetLogPriorTerm()Returns a value that is proportional to the log of the prior.doublegetLogProbAndPartialDerivationFor(boolean forward, int startPos, int endPos, IntList indices, DoubleList partDer, Sequence seq)doublegetLogProbFor(boolean forward, int startPos, int endPos, Sequence seq)This method computes the logarithm of the likelihood.StringgetNodeLabel(double weight, String name, NumberFormat nf)Returns the graphviz label of the node containing this emission.StringgetNodeShape(boolean forward)Returns the graphviz string for the shape of the node.intgetNumberOfParameters()Returns the number of parameters of this emission.intgetSizeOfEventSpace()Returns the size of the event space, i.e., the number of possible outcomes, for the random variables of this emissionvoidinitForSampling(int starts)This method initializes the instance for the sampling.voidinitializeFunctionRandomly()This method initializes the emission randomly.booleanisInSamplingMode()This method returnstrueif the object is currently used in a sampling, otherwisefalse.voidjoinStatistics(Emission... emissions)This method joins the statistics of different instances and sets this joined statistic as statistic of each instance.booleanparseNextParameterSet()This method allows the user to parse the next set of parameters (from a file).booleanparseParameterSet(int sampling, int n)This method allows the user to parse the set of parameters with indexnof a certainsampling(from a file).voidresetStatistic()This method resets the internal sufficient statistic.voidsamplingStopped()This method is the opposite of the methodSamplingComponent.extendSampling(int, boolean).voidsetParameter(double[] params, int offset)This method sets the internal parameters using the given global parameter array, the global offset of the HMM and the internal offset.intsetParameterOffset(int offset)This method sets the internal parameter offset and returns the new parameter offset for further use.voidsetParameters(Emission t)Set values of parameters of the instance to the value of the parameters of the given instance.StringtoString(NumberFormat nf)This method returns aStringrepresentation of the instance.StringBuffertoXML()This method returns an XML representation asStringBufferof an instance of the implementing class.
-
-
-
Constructor Detail
-
SilentEmission
public SilentEmission()
The main constructor.
-
SilentEmission
public SilentEmission(StringBuffer xml)
The standard constructor for the interfaceStorable. Constructs aSilentEmissionout of an XML representation.- Parameters:
xml- the XML representation asStringBuffer
-
-
Method Detail
-
clone
public SilentEmission clone() throws CloneNotSupportedException
- Overrides:
clonein classObject- Throws:
CloneNotSupportedException
-
toXML
public StringBuffer toXML()
Description copied from interface:StorableThis method returns an XML representation asStringBufferof an instance of the implementing class.
-
getLogProbAndPartialDerivationFor
public double getLogProbAndPartialDerivationFor(boolean forward, int startPos, int endPos, IntList indices, DoubleList partDer, Sequence seq) throws OperationNotSupportedExceptionDescription copied from interface:DifferentiableEmissionReturns the logarithmic score for aSequencebeginning at positionstartin theSequenceand fills lists with the indices and the partial derivations.- Specified by:
getLogProbAndPartialDerivationForin interfaceDifferentiableEmission- Parameters:
forward- a switch whether to use the forward or the reverse complementary strand of the sequencestartPos- the start position in theSequenceendPos- the end position in theSequenceindices- anIntListof indices, after method invocation the list should contain the indices i where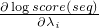 is not zero
is not zeropartDer- aDoubleListof partial derivations, after method invocation the list should contain the corresponding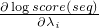 that are not zero
that are not zeroseq- theSequence- Returns:
- the logarithmic score for the
Sequence - Throws:
OperationNotSupportedException- ifforward==falseand the reverse complement of the sequence can not be computed
-
getLogProbFor
public double getLogProbFor(boolean forward, int startPos, int endPos, Sequence seq) throws OperationNotSupportedExceptionDescription copied from interface:EmissionThis method computes the logarithm of the likelihood.- Specified by:
getLogProbForin interfaceEmission- Parameters:
forward- whether to use the forward or the reverse strandstartPos- the start positionendPos- the end positionseq- the sequence- Returns:
- the logarithm of the probability
- Throws:
OperationNotSupportedException- ifforward=falseand the reverse complement of the sequenceseqis not defined
-
joinStatistics
public void joinStatistics(Emission... emissions)
Description copied from interface:EmissionThis method joins the statistics of different instances and sets this joined statistic as statistic of each instance. This method might be used for instance in a multi-threaded optimization to join partial statistics.- Specified by:
joinStatisticsin interfaceEmission- Parameters:
emissions- the emissions to be joined
-
addToStatistic
public void addToStatistic(boolean forward, int startPos, int endPos, double weight, Sequence seq) throws OperationNotSupportedExceptionDescription copied from interface:EmissionThis method adds theweightto the internal sufficient statistic.- Specified by:
addToStatisticin interfaceEmission- Parameters:
forward- whether to use the forward or the reverse strandstartPos- the start positionendPos- the end positionweight- the weight of the sequenceseq- the sequence- Throws:
OperationNotSupportedException- ifforward=falseand the reverse complement of the sequenceseqis not defined
-
estimateFromStatistic
public void estimateFromStatistic()
Description copied from interface:EmissionThis method estimates the parameters from the internal sufficient statistic.- Specified by:
estimateFromStatisticin interfaceEmission
-
resetStatistic
public void resetStatistic()
Description copied from interface:EmissionThis method resets the internal sufficient statistic.- Specified by:
resetStatisticin interfaceEmission
-
getAlphabetContainer
public AlphabetContainer getAlphabetContainer()
Description copied from interface:EmissionThis method returns theAlphabetContainerof this emission.- Specified by:
getAlphabetContainerin interfaceEmission- Returns:
- the
AlphabetContainerof this emission
-
addGradientOfLogPriorTerm
public void addGradientOfLogPriorTerm(double[] gradient, int offset)Description copied from interface:DifferentiableEmissionThis method computes the gradient ofEmission.getLogPriorTerm()for each parameter of this model. The results are added to the arraygradbeginning at index (offset+ internal offset).- Specified by:
addGradientOfLogPriorTermin interfaceDifferentiableEmission- Parameters:
gradient- the array of gradientsoffset- the start index of the HMM in thegradarray, where the partial derivations for the parameters of the HMM shall be entered- See Also:
Emission.getLogPriorTerm(),DifferentiableEmission.setParameterOffset(int)
-
getLogPriorTerm
public double getLogPriorTerm()
Description copied from interface:EmissionReturns a value that is proportional to the log of the prior. For maximum likelihood (ML) 0 should be returned.- Specified by:
getLogPriorTermin interfaceEmission- Returns:
- a value that is proportional to the log of the prior
- See Also:
StatisticalModel.getLogPriorTerm()
-
fillCurrentParameter
public void fillCurrentParameter(double[] params)
Description copied from interface:DifferentiableEmissionFills the current parameters in the globalparamsarray using the internal offset.- Specified by:
fillCurrentParameterin interfaceDifferentiableEmission- Parameters:
params- the global parameter array of the HMM- See Also:
DifferentiableEmission.setParameterOffset(int)
-
setParameter
public void setParameter(double[] params, int offset)Description copied from interface:DifferentiableEmissionThis method sets the internal parameters using the given global parameter array, the global offset of the HMM and the internal offset.- Specified by:
setParameterin interfaceDifferentiableEmission- Parameters:
params- the global parameter array of the classifieroffset- the offset of the HMM- See Also:
DifferentiableEmission.setParameterOffset(int)
-
setParameterOffset
public int setParameterOffset(int offset)
Description copied from interface:DifferentiableEmissionThis method sets the internal parameter offset and returns the new parameter offset for further use.- Specified by:
setParameterOffsetin interfaceDifferentiableEmission- Parameters:
offset- the offset to be set- Returns:
- the new parameter offset
-
initializeFunctionRandomly
public void initializeFunctionRandomly()
Description copied from interface:EmissionThis method initializes the emission randomly.- Specified by:
initializeFunctionRandomlyin interfaceEmission
-
drawParametersFromStatistic
public void drawParametersFromStatistic()
Description copied from interface:SamplingFromStatisticThis method draws the parameters using a sufficient statistic representing a posteriori density. It is recommended to write the parameters to a specific file usingSamplingComponent.acceptParameters()so that they can later be parsed using the methods of the interface.
Before using this method the methodSamplingComponent.initForSampling(int)should be called.- Specified by:
drawParametersFromStatisticin interfaceSamplingFromStatistic- See Also:
SamplingComponent.initForSampling(int),SamplingComponent.acceptParameters()
-
extendSampling
public void extendSampling(int sampling, boolean append) throws IOExceptionDescription copied from interface:SamplingComponentThis method allows to extend a sampling.- Specified by:
extendSamplingin interfaceSamplingComponent- Parameters:
sampling- the index of the samplingappend- whether to append the sampled parameters to an existing file or to overwrite the file- Throws:
IOException- if the file could not be handled correctly
-
initForSampling
public void initForSampling(int starts) throws IOExceptionDescription copied from interface:SamplingComponentThis method initializes the instance for the sampling. For instance this method can be used to create new files where all parameter sets are stored.- Specified by:
initForSamplingin interfaceSamplingComponent- Parameters:
starts- the number of different sampling starts that will be done- Throws:
IOException- if something went wrong- See Also:
File.createTempFile(String, String, java.io.File )
-
isInSamplingMode
public boolean isInSamplingMode()
Description copied from interface:SamplingComponentThis method returnstrueif the object is currently used in a sampling, otherwisefalse.- Specified by:
isInSamplingModein interfaceSamplingComponent- Returns:
trueif the object is currently used in a sampling, otherwisefalse
-
parseNextParameterSet
public boolean parseNextParameterSet()
Description copied from interface:SamplingComponentThis method allows the user to parse the next set of parameters (from a file).- Specified by:
parseNextParameterSetin interfaceSamplingComponent- Returns:
trueif the parameters could be parsed, otherwisefalse- See Also:
SamplingComponent.parseParameterSet(int, int)
-
parseParameterSet
public boolean parseParameterSet(int sampling, int n) throws ExceptionDescription copied from interface:SamplingComponentThis method allows the user to parse the set of parameters with indexnof a certainsampling(from a file). The internal numbering should start with 0. The parameter set with index 0 is the initial (random) parameter set. It is recommended that a series of parameter sets is accessed by the following lines:for( sampling = 0; sampling < numSampling; sampling++ )
{
boolean b = parseParameterSet( sampling, n );
while( b )
{
//do something
b = parseNextParameterSet();
}
}
- Specified by:
parseParameterSetin interfaceSamplingComponent- Parameters:
sampling- the index of the samplingn- the index of the parameter set- Returns:
trueif the parameter set could be parsed- Throws:
Exception- if there is a problem with parsing the parameters- See Also:
SamplingComponent.parseNextParameterSet()
-
samplingStopped
public void samplingStopped() throws IOExceptionDescription copied from interface:SamplingComponentThis method is the opposite of the methodSamplingComponent.extendSampling(int, boolean). It can be used for closing any streams of writer, ...- Specified by:
samplingStoppedin interfaceSamplingComponent- Throws:
IOException- if something went wrong- See Also:
SamplingComponent.extendSampling(int, boolean)
-
acceptParameters
public void acceptParameters() throws IOExceptionDescription copied from interface:SamplingComponentThis methods accepts the drawn parameters. Internally the drawn parameters should be saved (to a file).- Specified by:
acceptParametersin interfaceSamplingComponent- Throws:
IOException- if the file could not be handled correctly
-
getLogPosteriorFromStatistic
public double getLogPosteriorFromStatistic()
Description copied from interface:SamplingFromStatisticThis method calculates the a-posteriori probability for the current statistics- Specified by:
getLogPosteriorFromStatisticin interfaceSamplingFromStatistic- Returns:
- the logarithm of the a-posteriori probability
-
toString
public String toString(NumberFormat nf)
Description copied from interface:EmissionThis method returns aStringrepresentation of the instance.- Specified by:
toStringin interfaceEmission- Parameters:
nf- theNumberFormatfor theStringrepresentation of parameters or probabilities- Returns:
- a
Stringrepresentation of the instance
-
getLogGammaScoreFromStatistic
public double getLogGammaScoreFromStatistic()
Description copied from interface:SamplingEmissionThis method calculates a score for the current statistics, which is independent from the current parameters In general the gamma-score is a product of gamma-functions parameterized with the current statistics- Specified by:
getLogGammaScoreFromStatisticin interfaceSamplingEmission- Returns:
- the logarithm of the gamma-score for the current statistics
-
getNodeShape
public String getNodeShape(boolean forward)
Description copied from interface:EmissionReturns the graphviz string for the shape of the node.- Specified by:
getNodeShapein interfaceEmission- Parameters:
forward- if this emission is used on the forward strand- Returns:
- the shape
-
getNodeLabel
public String getNodeLabel(double weight, String name, NumberFormat nf)
Description copied from interface:EmissionReturns the graphviz label of the node containing this emission.- Specified by:
getNodeLabelin interfaceEmission- Parameters:
weight- the weight of the node which is represented by the color of the node, or -1 for no representation, i.e., white backgroundname- the name of the state using this emissionnf- theNumberFormatfor formatting the textual representation of this emission- Returns:
- the label
-
fillSamplingGroups
public void fillSamplingGroups(int parameterOffset, LinkedList<int[]> list)Description copied from interface:DifferentiableEmissionAdds the groups of indexes of those parameters of this emission that should be sampled together in one step of a grouped sampling procedure, each as anint[], intolist. In most cases, one group should contain the parameters that are living on a common simplex. The internal indexes of the parameters are incremeneted by an externalparameterOffset- Specified by:
fillSamplingGroupsin interfaceDifferentiableEmission- Parameters:
parameterOffset- the external parameter offsetlist- the list of sampling groups
-
getNumberOfParameters
public int getNumberOfParameters()
Description copied from interface:DifferentiableEmissionReturns the number of parameters of this emission.- Specified by:
getNumberOfParametersin interfaceDifferentiableEmission- Returns:
- the number of parameters
-
getSizeOfEventSpace
public int getSizeOfEventSpace()
Description copied from interface:DifferentiableEmissionReturns the size of the event space, i.e., the number of possible outcomes, for the random variables of this emission- Specified by:
getSizeOfEventSpacein interfaceDifferentiableEmission- Returns:
- the size of the event space
-
setParameters
public void setParameters(Emission t) throws IllegalArgumentException
Description copied from interface:EmissionSet values of parameters of the instance to the value of the parameters of the given instance. It can be assumed that the given instance and the current instance are from the same class. This method might be used for instance in a multi-threaded optimization to broadcast the parameters.- Specified by:
setParametersin interfaceEmission- Parameters:
t- the emission with the parameters to be set- Throws:
IllegalArgumentException- if the assumption about the same class for given and current instance is wrong
-
-