Cookbook
You can also download a PDF version of this cookbook.
Sequence analysis is one of the major subjects of bioinformatics. Several existing libraries combine the representation of biological sequences with exact and approximate pattern matching as well as alignment algorithms. We present Jstacs, an open source Java library, which focuses on the statistical analysis of biological sequences instead. Jstacs comprises an efficient representation of sequence data and provides implementations of many statistical models with generative and discriminative approaches for parameter learning. Using Jstacs, classifiers can be assessed and compared on test datasets or by cross-validation experiments evaluating several performance measures. Due to its strictly object-oriented design Jstacs is easy to use and readily extensible.
Jstacs is a joint project of the groups Bioinformatics and Pattern Recognition and Bioinformatics at the Institute of Computer Science of Martin Luther University Halle-Wittenberg and the Research Group Data Inspection at the Leibniz Institute of Plant Genetics and Crop Plant Research.
Jstacs is listed in the machine learning open-source software (mloss) repository.
The complete API documentation can be found at http://www.jstacs.de/api-2.1/.
A reference card of Jstacs classes and methods can be found as a separate download.
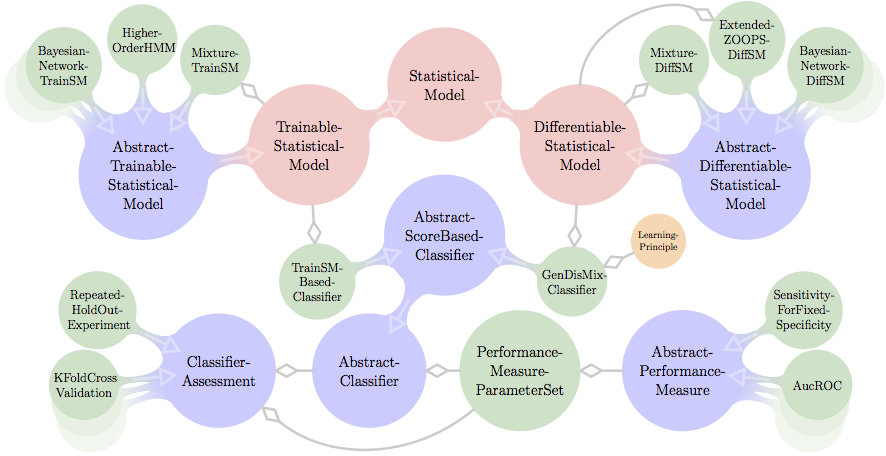
General structure of Jstacs
A coarse view on the structure of Jstacs is presented in Figure below. Being a library for statistical analysis and classification of sequence data, Jstacs is organized around the abstract class AbstractClassifier, the interface StatisticalModel and its two sub-interfaces TrainableStatisticalModel, and DifferentiableStatisticalModel.
StatisticalModel s represent statistical models in general, which can compute the log-likelihood of a given input sequence and define prior densities on their parameters. The abstract implementation AbstractTrainSM of TrainableStatisticalModel is the base class of many generatively learned models such as Bayesian networks, hidden Markov models, or mixture models, and can be learned from a single input data set. In constrast, DifferentiableStatisticalModel s provide all facilities for numerical optimization of parameters, which is especially necessary for discriminative parameter learning. The abstract base class of all DifferentiableStatisticalModel implementations is AbstractDifferentiableStatisticalModel. Currently, DifferentiableStatisticalModel s include Bayesian networks, Markov models, a ZOOPS model, and mixture models.
AbstractClassifier defines the general properties of a classifier. Its sub-class AbstractScoreBasedClassifier adds additional methods for the classification of sequences based on a sequence and class-specific score. Two concrete sub-classes of AbstractScoreBasedClassifier are the TrainSMBasedClassifier, which works on TrainableStatisticalModel s, and the GenDisMixClassifier, which works on DifferentiableStatisticalModel.
AbstractClassifier s can be assessed either on dedicated training and test data sets or in ClassifierAssessment s like KFoldCrossValidation or RepeatedHoldOutExperiment. The performance measures used in such an assessment are collected in PerformanceMeasureParameterSet s containing sub-classes of the abstract class AbstractPerformanceMeasure.
A more detailed view on all of these classes will be given in the remainder of this cookbook.
About this cookbook
This document is not a cookbook in the sense of a collection of recipes. The intention of this cookbook is rather to learn how to cook with the ingredients and tools provided by Jstacs.
Nonetheless, we present a collection of recipes in the last section, where you find the code of executable code examples that can also be downloaded as a zip file.
We are aware that a library of this size seems daunting on first sight -- and on second sight as well. However, we hope that despite the inevitable complexity and size of such a library, this cookbook may help to get a picture of the structure, design principles, and capabilities of Jstacs.
This cookbook is structured as follows: in the section Quick start: Jstacs in a nutshell, we give a very brief introduction to Jstacs enabling to run first programs without reading the complete cookbook.
In section Starter: Data handling, we explain how data are represented in Jstacs, and how you can read data from files.
In section Intermediate course: XMLParser, Parameters, and Results, we present some facilities, an XML parser and the representation of parameters and results, that are used frequently within Jstacs and are necessary for the following parts.
In section First main course: SequenceScores, we present sequence scores, statistical models, and their sub-interfaces and sub-classes. We explain the methods defined in these interfaces and classes, and we show how you can create and use their existing implementations.
In section Second main course: Classifiers, we explain classifiers and assessment of classifiers using different performance measures.
In section Intermediate course: Optimization we present the facilities for numerical optimization.
In section Dessert: Alignments, Utils, and goodies, we list utility classes and methods, that we think might be of help for your own implementations.
Finally, in section Recipes, we give a number of executable code examples that may serve as a starting point of your own classes and applications.