de.jstacs.classifier.scoringFunctionBased.gendismix
Class LogGenDisMixFunction
java.lang.Object
 de.jstacs.algorithms.optimization.DifferentiableFunction
de.jstacs.algorithms.optimization.DifferentiableFunction
 de.jstacs.classifier.scoringFunctionBased.OptimizableFunction
de.jstacs.classifier.scoringFunctionBased.OptimizableFunction
 de.jstacs.classifier.scoringFunctionBased.AbstractOptimizableFunction
de.jstacs.classifier.scoringFunctionBased.AbstractOptimizableFunction
 de.jstacs.classifier.scoringFunctionBased.AbstractMultiThreadedOptimizableFunction
de.jstacs.classifier.scoringFunctionBased.AbstractMultiThreadedOptimizableFunction
 de.jstacs.classifier.scoringFunctionBased.SFBasedOptimizableFunction
de.jstacs.classifier.scoringFunctionBased.SFBasedOptimizableFunction
 de.jstacs.classifier.scoringFunctionBased.gendismix.LogGenDisMixFunction
de.jstacs.classifier.scoringFunctionBased.gendismix.LogGenDisMixFunction
- All Implemented Interfaces:
- Function
- Direct Known Subclasses:
- OneSampleLogGenDisMixFunction
public class LogGenDisMixFunction
- extends SFBasedOptimizableFunction
This class implements the the following function
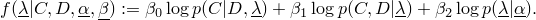
The weights 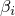 have to sum to 1. For special weights the optimization turns out to be
well known
have to sum to 1. For special weights the optimization turns out to be
well known
- if the weights are (0,1,0), one obtains maximum likelihood,
- if the weights are (0,0.5,0.5), one obtains maximum a posteriori,
- if the weights are (1,0,0), one obtains maximum conditional likelihood,
- if the weights are (0.5,0,0.5), one obtains maximum supervised posterior,
- if the
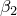 =0, one obtains the generative-discriminative trade-off,
=0, one obtains the generative-discriminative trade-off,
- if the
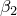 =0.5, one obtains the penalized generative-discriminative trade-off.
=0.5, one obtains the penalized generative-discriminative trade-off.
Of course, there are also some very interesting cases with other weights.
It can be used to maximize the parameters.
This class enables the user to exploit all CPUs of the computer by using threads. The number of compute threads can be
determined in the constructor.
It is very important for this class that the ScoringFunction.clone() method works correctly, since each thread works on its own clones.
- Author:
- Jens Keilwagen
|
Field Summary |
protected double[] |
beta
The mixture parameters of the GenDisMix |
protected double[][] |
cllGrad
Array for the gradient of the conditional log-likelihood |
protected double[][] |
helpArray
General temporary array |
protected double[][] |
llGrad
Array for the gradient of the log-likelihood |
protected double[] |
prGrad
Array for the gradient of the prior |
|
Method Summary |
protected void |
evaluateFunction(int index,
int startClass,
int startSeq,
int endClass,
int endSeq)
This method evaluates the function for a part of the data. |
protected void |
evaluateGradientOfFunction(int index,
int startClass,
int startSeq,
int endClass,
int endSeq)
This method evaluates the gradient of the function for a part of the data. |
protected double |
joinFunction()
This method joins the partial results that have been computed using AbstractMultiThreadedOptimizableFunction.evaluateFunction(int, int, int, int, int). |
protected double[] |
joinGradients()
This method joins the gradients of each part that have been computed using AbstractMultiThreadedOptimizableFunction.evaluateGradientOfFunction(int, int, int, int, int). |
void |
reset(ScoringFunction[] funs)
This method allows to reset the internally used functions and the corresponding objects. |
| Methods inherited from class java.lang.Object |
clone, equals, finalize, getClass, hashCode, notify, notifyAll, toString, wait, wait, wait |
helpArray
protected double[][] helpArray
- General temporary array
llGrad
protected double[][] llGrad
- Array for the gradient of the log-likelihood
cllGrad
protected double[][] cllGrad
- Array for the gradient of the conditional log-likelihood
beta
protected double[] beta
- The mixture parameters of the GenDisMix
prGrad
protected double[] prGrad
- Array for the gradient of the prior
LogGenDisMixFunction
public LogGenDisMixFunction(int threads,
ScoringFunction[] score,
Sample[] data,
double[][] weights,
LogPrior prior,
double[] beta,
boolean norm,
boolean freeParams)
throws IllegalArgumentException
- The constructor for creating an instance that can be used in an
Optimizer.
- Parameters:
threads - the number of threads used for evaluating the function and determining the gradient of the functionscore - an array containing the ScoringFunctions that are used for determining the sequences scores;
if the weight beta[LearningPrinciple.LIKELIHOOD_INDEX] is positive all elements of score have to be NormalizableScoringFunctiondata - the array of Samples containing the data that is needed to evaluate the functionweights - the weights for each Sequence in each Sample of dataprior - the prior that is used for learning the parametersbeta - the beta-weights for the three terms of the learning principlenorm - the switch for using the normalization (division by the number
of sequences)freeParams - the switch for using only the free parameters
- Throws:
IllegalArgumentException - if the number of threads is not positive, the number of classes or the dimension of the weights is not correct
joinGradients
protected double[] joinGradients()
throws EvaluationException
- Description copied from class:
AbstractMultiThreadedOptimizableFunction
- This method joins the gradients of each part that have been computed using
AbstractMultiThreadedOptimizableFunction.evaluateGradientOfFunction(int, int, int, int, int).
- Specified by:
joinGradients in class AbstractMultiThreadedOptimizableFunction
- Returns:
- the gradient
- Throws:
EvaluationException - if the gradient could not be evaluated properly
evaluateGradientOfFunction
protected void evaluateGradientOfFunction(int index,
int startClass,
int startSeq,
int endClass,
int endSeq)
- Description copied from class:
AbstractMultiThreadedOptimizableFunction
- This method evaluates the gradient of the function for a part of the data.
- Specified by:
evaluateGradientOfFunction in class AbstractMultiThreadedOptimizableFunction
- Parameters:
index - the index of the partstartClass - the index of the start classstartSeq - the index of the start sequenceendClass - the index of the end class (inclusive)endSeq - the index of the end sequence (exclusive)
joinFunction
protected double joinFunction()
throws DimensionException,
EvaluationException
- Description copied from class:
AbstractMultiThreadedOptimizableFunction
- This method joins the partial results that have been computed using
AbstractMultiThreadedOptimizableFunction.evaluateFunction(int, int, int, int, int).
- Specified by:
joinFunction in class AbstractMultiThreadedOptimizableFunction
- Returns:
- the value of the function
- Throws:
DimensionException - if the parameters could not be set
EvaluationException - if the gradient could not be evaluated properly
evaluateFunction
protected void evaluateFunction(int index,
int startClass,
int startSeq,
int endClass,
int endSeq)
throws EvaluationException
- Description copied from class:
AbstractMultiThreadedOptimizableFunction
- This method evaluates the function for a part of the data.
- Specified by:
evaluateFunction in class AbstractMultiThreadedOptimizableFunction
- Parameters:
index - the index of the partstartClass - the index of the start classstartSeq - the index of the start sequenceendClass - the index of the end class (inclusive)endSeq - the index of the end sequence (exclusive)
- Throws:
EvaluationException - if the gradient could not be evaluated properly
reset
public void reset(ScoringFunction[] funs)
throws Exception
- Description copied from class:
SFBasedOptimizableFunction
- This method allows to reset the internally used functions and the corresponding objects.
- Specified by:
reset in class SFBasedOptimizableFunction
- Parameters:
funs - the new instances
- Throws:
Exception - if something went wrong
 de.jstacs.algorithms.optimization.DifferentiableFunction
de.jstacs.algorithms.optimization.DifferentiableFunction
 de.jstacs.classifier.scoringFunctionBased.OptimizableFunction
de.jstacs.classifier.scoringFunctionBased.OptimizableFunction
 de.jstacs.classifier.scoringFunctionBased.AbstractOptimizableFunction
de.jstacs.classifier.scoringFunctionBased.AbstractOptimizableFunction
 de.jstacs.classifier.scoringFunctionBased.AbstractMultiThreadedOptimizableFunction
de.jstacs.classifier.scoringFunctionBased.AbstractMultiThreadedOptimizableFunction
 de.jstacs.classifier.scoringFunctionBased.SFBasedOptimizableFunction
de.jstacs.classifier.scoringFunctionBased.SFBasedOptimizableFunction
 de.jstacs.classifier.scoringFunctionBased.gendismix.LogGenDisMixFunction
de.jstacs.classifier.scoringFunctionBased.gendismix.LogGenDisMixFunction
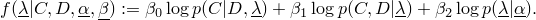
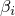 have to sum to 1. For special weights the optimization turns out to be
well known
have to sum to 1. For special weights the optimization turns out to be
well known
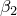 =0, one obtains the generative-discriminative trade-off,
=0, one obtains the generative-discriminative trade-off,