- java.lang.Object
-
- de.jstacs.sequenceScores.differentiable.AbstractDifferentiableSequenceScore
-
- de.jstacs.sequenceScores.statisticalModels.differentiable.AbstractDifferentiableStatisticalModel
-
- de.jstacs.sequenceScores.statisticalModels.differentiable.directedGraphicalModels.BayesianNetworkDiffSM
-
- All Implemented Interfaces:
- InstantiableFromParameterSet, DifferentiableSequenceScore, QuickScanningSequenceScore, SequenceScore, DifferentiableStatisticalModel, StatisticalModel, Storable, Cloneable
- Direct Known Subclasses:
- MarkovModelDiffSM
public class BayesianNetworkDiffSM extends AbstractDifferentiableStatisticalModel implements InstantiableFromParameterSet, QuickScanningSequenceScore
This class implements a scoring function that is a moral directed graphical model, i.e. a moral Bayesian network. This implementation also comprises well known specializations of Bayesian networks like Markov models of arbitrary order (including weight array matrix models (WAM) and position weight matrices (PWM)) or Bayesian trees. Different structures can be achieved by using the correspondingMeasure, e.g.InhomogeneousMarkovfor Markov models of arbitrary order.
This scoring function can be used in anyScoreClassifier, e.g. in aMSPClassifierto learn the parameters of theDifferentiableStatisticalModelusing maximum conditional likelihood or maximum supervised posterior.- Author:
- Jan Grau
-
-
Field Summary
Fields Modifier and Type Field and Description protected doubleessThe equivalent sample size.protected booleanisTrainedIndicates if the instance has been trained.protected DoublelogNormalizationConstantNormalization constant to obtain normalized probabilities.protected IntegernumFreeParsThe number of free parameters.protected int[]numsUsed internally.protected int[][]orderThe network structure, used internally.protected BNDiffSMParameter[]parametersThe parameters of the scoring function.protected booleanplugInParametersIndicates if plug-in parameters, i.e.protected MeasurestructureMeasureMeasurethat defines the network structure.protected BNDiffSMParameterTree[]treesThe trees that represent the context of the random variable (i.e.-
Fields inherited from class de.jstacs.sequenceScores.differentiable.AbstractDifferentiableSequenceScore
alphabets, length, r
-
Fields inherited from interface de.jstacs.sequenceScores.differentiable.DifferentiableSequenceScore
UNKNOWN
-
-
Constructor Summary
Constructors Constructor and Description BayesianNetworkDiffSM(AlphabetContainer alphabet, int length, double ess, boolean plugInParameters, Measure structureMeasure)Creates a newBayesianNetworkDiffSMthat has neither been initialized nor trained.BayesianNetworkDiffSM(BayesianNetworkDiffSMParameterSet parameters)Creates a newBayesianNetworkDiffSMthat has neither been initialized nor trained from aBayesianNetworkDiffSMParameterSet.BayesianNetworkDiffSM(StringBuffer xml)The standard constructor for the interfaceStorable.
-
Method Summary
All Methods Instance Methods Concrete Methods Modifier and Type Method and Description voidaddGradientOfLogPriorTerm(double[] grad, int start)This method computes the gradient ofDifferentiableStatisticalModel.getLogPriorTerm()for each parameter of this model.BayesianNetworkDiffSMclone()Creates a clone (deep copy) of the currentDifferentiableSequenceScoreinstance.protected voidcreateTrees(DataSet[] data2, double[][] weights2)Creates the tree structures that represent the context (arraytrees) and the parameter objectsparametersusing the givenMeasurestructureMeasure.DataSetemitDataSet(int numberOfSequences, int... seqLength)This method returns aDataSetobject containing artificial sequence(s).voidfillInfixScore(int[] seq, int start, int length, double[] scores)Computes the position-wise scores of an infix of the sequenceseq(which must be encoded by the same alphabet as thisQuickScanningSequenceScore) beginning atstartand extending forlengthpositions.protected voidfromXML(StringBuffer source)This method is called in the constructor for theStorableinterface to create a scoring function from aStringBuffer.InstanceParameterSetgetCurrentParameterSet()Returns theInstanceParameterSetthat has been used to instantiate the current instance of the implementing class.double[]getCurrentParameterValues()Returns adoublearray of dimensionDifferentiableSequenceScore.getNumberOfParameters()containing the current parameter values.doublegetESS()Returns the equivalent sample size (ess) of this model, i.e.boolean[][]getInfixFilter(int kmer, double thresh, int... start)Computes arrays that indicate, for a given set of starting positions and a given k-mer length, if a sequence containing this k-mer may yield a score abovethreshold, choosing the best-scoring option among all non-specified positions (i.e., those outside the k-mer).StringgetInstanceName()Should return a short instance name such as iMM(0), BN(2), ...doublegetLogNormalizationConstant()Returns the logarithm of the sum of the scores over all sequences of the event space.doublegetLogPartialNormalizationConstant(int parameterIndex)Returns the logarithm of the partial normalization constant for the parameter with indexparameterIndex.doublegetLogPriorTerm()This method computes a value that is proportional todoublegetLogScoreAndPartialDerivation(Sequence seq, int start, IntList indices, DoubleList partialDer)doublegetLogScoreFor(Sequence seq, int start)bytegetMaximalMarkovOrder()This method returns the maximal used Markov order, if possible.doublegetMaximumScore()Returns the maximum score of thisBayesianNetworkDiffSMreturned for a admissible input sequence.intgetNumberOfParameters()Returns the number of parameters in thisDifferentiableSequenceScore.double[]getPositionDependentKMerProb(Sequence kmer)Returns the probability ofkmerfor all possible positions in thisBayesianNetworkDiffSMstarting at positionkmer.getLength()-1.intgetPositionForParameter(int index)Returns the position in the sequence the parameterindexis responsible for.double[][]getPWM()If thisBayesianNetworkDiffSMis a PWM, i.e.intgetSizeOfEventSpaceForRandomVariablesOfParameter(int index)Returns the size of the event space of the random variables that are affected by parameter no.voidinitializeFunction(int index, boolean freeParams, DataSet[] data, double[][] weights)This method creates the underlying structure of theDifferentiableSequenceScore.voidinitializeFunctionRandomly(boolean freeParams)This method initializes theDifferentiableSequenceScorerandomly.booleanisInitialized()This method can be used to determine whether the instance is initialized.protected voidprecomputeNormalization()Pre-computes all normalization constants.voidsetParameters(double[] params, int start)This method sets the internal parameters to the values ofparamsbetweenstartandstart +DifferentiableSequenceScore.getNumberOfParameters()- 1protected voidsetPlugInParameters(int index, boolean freeParameters, DataSet[] data, double[][] weights)Computes and sets the plug-in parameters (MAP estimated parameters) fromdatausingweights.StringtoHtml(NumberFormat nf)Returns an HTML representation of thisBayesianNetworkDiffSM.StringtoString(NumberFormat nf)This method returns aStringrepresentation of the instance.StringBuffertoXML()This method returns an XML representation asStringBufferof an instance of the implementing class.-
Methods inherited from class de.jstacs.sequenceScores.statisticalModels.differentiable.AbstractDifferentiableStatisticalModel
getInitialClassParam, getLogProbFor, getLogProbFor, getLogProbFor, getLogScoreFor, getLogScoreFor, isNormalized, isNormalized
-
Methods inherited from class de.jstacs.sequenceScores.differentiable.AbstractDifferentiableSequenceScore
getAlphabetContainer, getCharacteristics, getLength, getLogScoreAndPartialDerivation, getLogScoreAndPartialDerivation, getLogScoreFor, getLogScoreFor, getNumberOfRecommendedStarts, getNumberOfStarts, getNumericalCharacteristics, toString
-
Methods inherited from class java.lang.Object
equals, finalize, getClass, hashCode, notify, notifyAll, wait, wait, wait
-
Methods inherited from interface de.jstacs.sequenceScores.SequenceScore
getAlphabetContainer, getCharacteristics, getLength, getLogScoreFor, getLogScoreFor, getLogScoreFor, getLogScoreFor, getNumericalCharacteristics
-
Methods inherited from interface de.jstacs.sequenceScores.differentiable.DifferentiableSequenceScore
getLogScoreAndPartialDerivation, getLogScoreAndPartialDerivation, getNumberOfRecommendedStarts
-
-
-
-
Field Detail
-
parameters
protected BNDiffSMParameter[] parameters
The parameters of the scoring function. This comprises free as well as dependent parameters.
-
trees
protected BNDiffSMParameterTree[] trees
The trees that represent the context of the random variable (i.e. configuration of parent random variables) of the parameters.
-
isTrained
protected boolean isTrained
Indicates if the instance has been trained.
-
ess
protected double ess
The equivalent sample size.
-
numFreePars
protected Integer numFreePars
The number of free parameters.
-
nums
protected int[] nums
Used internally. Mapping from indexes of free parameters to indexes of all parameters.
-
plugInParameters
protected boolean plugInParameters
Indicates if plug-in parameters, i.e. generative (MAP) parameters shall be used upon initialization.
-
order
protected int[][] order
The network structure, used internally.
-
logNormalizationConstant
protected Double logNormalizationConstant
Normalization constant to obtain normalized probabilities.
-
-
Constructor Detail
-
BayesianNetworkDiffSM
public BayesianNetworkDiffSM(AlphabetContainer alphabet, int length, double ess, boolean plugInParameters, Measure structureMeasure) throws Exception
Creates a newBayesianNetworkDiffSMthat has neither been initialized nor trained.- Parameters:
alphabet- the alphabet of the scoring function boxed in anAlphabetContainer, e.gnew AlphabetContainer(new DNAAlphabet())length- the length of the scoring function, i.e. the length of the sequences this scoring function can handleess- the equivalent sample sizeplugInParameters- indicates if plug-in parameters, i.e. generative (MAP) parameters, shall be used upon initializationstructureMeasure- theMeasureused for the structure, e.g.InhomogeneousMarkov- Throws:
Exception- if the length of the scoring function is not admissible (<=0) or the alphabet is not discrete
-
BayesianNetworkDiffSM
public BayesianNetworkDiffSM(BayesianNetworkDiffSMParameterSet parameters) throws ParameterSetParser.NotInstantiableException, Exception
Creates a newBayesianNetworkDiffSMthat has neither been initialized nor trained from aBayesianNetworkDiffSMParameterSet.- Parameters:
parameters- the parameter set- Throws:
ParameterSetParser.NotInstantiableException- if theBayesianNetworkDiffSMcould not be instantiated from theBayesianNetworkDiffSMParameterSetException- if the length of the scoring function is not admissible (<=0) or the alphabet is not discrete
-
BayesianNetworkDiffSM
public BayesianNetworkDiffSM(StringBuffer xml) throws NonParsableException
The standard constructor for the interfaceStorable. Recreates aBayesianNetworkDiffSMfrom its XML representation as saved by the methodtoXML().- Parameters:
xml- the XML representation asStringBuffer- Throws:
NonParsableException- if the XML code could not be parsed
-
-
Method Detail
-
clone
public BayesianNetworkDiffSM clone() throws CloneNotSupportedException
Description copied from interface:DifferentiableSequenceScoreCreates a clone (deep copy) of the currentDifferentiableSequenceScoreinstance.- Specified by:
clonein interfaceDifferentiableSequenceScore- Specified by:
clonein interfaceSequenceScore- Overrides:
clonein classAbstractDifferentiableStatisticalModel- Returns:
- the cloned instance of the current
DifferentiableSequenceScore - Throws:
CloneNotSupportedException- if something went wrong while cloning theDifferentiableSequenceScore
-
getLogPartialNormalizationConstant
public double getLogPartialNormalizationConstant(int parameterIndex) throws ExceptionDescription copied from interface:DifferentiableStatisticalModelReturns the logarithm of the partial normalization constant for the parameter with indexparameterIndex. This is the logarithm of the partial derivation of the normalization constant for the parameter with indexparameterIndex,- Specified by:
getLogPartialNormalizationConstantin interfaceDifferentiableStatisticalModel- Parameters:
parameterIndex- the index of the parameter- Returns:
- the logarithm of the partial normalization constant
- Throws:
Exception- if something went wrong with the normalization- See Also:
DifferentiableStatisticalModel.getLogNormalizationConstant()
-
initializeFunction
public void initializeFunction(int index, boolean freeParams, DataSet[] data, double[][] weights) throws ExceptionDescription copied from interface:DifferentiableSequenceScoreThis method creates the underlying structure of theDifferentiableSequenceScore.- Specified by:
initializeFunctionin interfaceDifferentiableSequenceScore- Parameters:
index- the index of the class theDifferentiableSequenceScoremodelsfreeParams- indicates whether the (reduced) parameterization is useddata- the data setsweights- the weights of the sequences in the data sets- Throws:
Exception- if something went wrong
-
createTrees
protected void createTrees(DataSet[] data2, double[][] weights2) throws Exception
Creates the tree structures that represent the context (arraytrees) and the parameter objectsparametersusing the givenMeasurestructureMeasure.- Parameters:
data2- the data that is used to compute the structureweights2- the weights on the sequences indata2- Throws:
Exception- if the structure is no moral graph or if the lengths of data and scoring function do not match or other problems concerning the data occur
-
setPlugInParameters
protected void setPlugInParameters(int index, boolean freeParameters, DataSet[] data, double[][] weights)Computes and sets the plug-in parameters (MAP estimated parameters) fromdatausingweights.- Parameters:
index- the index of the class the scoring function is responsible for, the parameters are estimated fromdata[index]andweights[index]freeParameters- indicates if only the free parameters or all parameters should be used, this also affects the initializationdata- the data used for initializationweights- the weights on the data
-
fromXML
protected void fromXML(StringBuffer source) throws NonParsableException
Description copied from class:AbstractDifferentiableSequenceScoreThis method is called in the constructor for theStorableinterface to create a scoring function from aStringBuffer.- Specified by:
fromXMLin classAbstractDifferentiableSequenceScore- Parameters:
source- the XML representation asStringBuffer- Throws:
NonParsableException- if theStringBuffercould not be parsed- See Also:
AbstractDifferentiableSequenceScore.AbstractDifferentiableSequenceScore(StringBuffer)
-
toString
public String toString(NumberFormat nf)
Description copied from interface:SequenceScoreThis method returns aStringrepresentation of the instance.- Specified by:
toStringin interfaceSequenceScore- Parameters:
nf- theNumberFormatfor theStringrepresentation of parameters or probabilities- Returns:
- a
Stringrepresentation of the instance
-
getInstanceName
public String getInstanceName()
Description copied from interface:SequenceScoreShould return a short instance name such as iMM(0), BN(2), ...- Specified by:
getInstanceNamein interfaceSequenceScore- Returns:
- a short instance name
-
getLogScoreFor
public double getLogScoreFor(Sequence seq, int start)
Description copied from interface:SequenceScore- Specified by:
getLogScoreForin interfaceSequenceScore- Parameters:
seq- theSequencestart- the start position in theSequence- Returns:
- the logarithmic score for the
Sequence
-
getInfixFilter
public boolean[][] getInfixFilter(int kmer, double thresh, int... start)Description copied from interface:QuickScanningSequenceScoreComputes arrays that indicate, for a given set of starting positions and a given k-mer length, if a sequence containing this k-mer may yield a score abovethreshold, choosing the best-scoring option among all non-specified positions (i.e., those outside the k-mer). This method is implemented as an upper bound on the scores, i.e., there may be k-mers that are considered to score above threshold (i.e., that have entrytrue) although they are not, but there may not be k-mers that are considered to be below threshold (i.e., that have entryfalse), although there exist sequences containing this k-mer that do. The returned array is indexed by the starting positions (in the same order as provided instarts) in the first dimension, and in the second dimension it is indexed by an integer representation of the k-mers, assigning the highest priority to the first k-mer position, i.e.,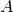 denotes the size of the alphabet,
denotes the size of the alphabet, 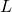 the length of the k-mer (starting at 0 in this case),
and
the length of the k-mer (starting at 0 in this case),
and 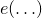 denotes the function encoding symbols from the alphabet as integers (see
denotes the function encoding symbols from the alphabet as integers (see DiscreteAlphabet).- Specified by:
getInfixFilterin interfaceQuickScanningSequenceScore- Parameters:
kmer- the k-mer lengththresh- the thresholdstart- the starting position(s)- Returns:
- the indicator arrays
-
getLogScoreAndPartialDerivation
public double getLogScoreAndPartialDerivation(Sequence seq, int start, IntList indices, DoubleList partialDer)
Description copied from interface:DifferentiableSequenceScoreReturns the logarithmic score for aSequencebeginning at positionstartin theSequenceand fills lists with the indices and the partial derivations.- Specified by:
getLogScoreAndPartialDerivationin interfaceDifferentiableSequenceScore- Parameters:
seq- theSequencestart- the start position in theSequenceindices- anIntListof indices, after method invocation the list should contain the indices i where is not zero
is not zeropartialDer- aDoubleListof partial derivations, after method invocation the list should contain the corresponding that are not zero
that are not zero- Returns:
- the logarithmic score for the
Sequence
-
getLogNormalizationConstant
public double getLogNormalizationConstant() throws RuntimeExceptionDescription copied from interface:DifferentiableStatisticalModelReturns the logarithm of the sum of the scores over all sequences of the event space.- Specified by:
getLogNormalizationConstantin interfaceDifferentiableStatisticalModel- Returns:
- the logarithm of the normalization constant Z
- Throws:
RuntimeException
-
getNumberOfParameters
public int getNumberOfParameters()
Description copied from interface:DifferentiableSequenceScoreReturns the number of parameters in thisDifferentiableSequenceScore. If the number of parameters is not known yet, the method returnsDifferentiableSequenceScore.UNKNOWN.- Specified by:
getNumberOfParametersin interfaceDifferentiableSequenceScore- Returns:
- the number of parameters in this
DifferentiableSequenceScore - See Also:
DifferentiableSequenceScore.UNKNOWN
-
setParameters
public void setParameters(double[] params, int start)Description copied from interface:DifferentiableSequenceScoreThis method sets the internal parameters to the values ofparamsbetweenstartandstart +DifferentiableSequenceScore.getNumberOfParameters()- 1- Specified by:
setParametersin interfaceDifferentiableSequenceScore- Parameters:
params- the new parametersstart- the start index inparams
-
precomputeNormalization
protected void precomputeNormalization()
Pre-computes all normalization constants.
-
getCurrentParameterValues
public double[] getCurrentParameterValues() throws ExceptionDescription copied from interface:DifferentiableSequenceScoreReturns adoublearray of dimensionDifferentiableSequenceScore.getNumberOfParameters()containing the current parameter values. If one likes to use these parameters to start an optimization it is highly recommended to invokeDifferentiableSequenceScore.initializeFunction(int, boolean, DataSet[], double[][])before. After an optimization this method can be used to get the current parameter values.- Specified by:
getCurrentParameterValuesin interfaceDifferentiableSequenceScore- Returns:
- the current parameter values
- Throws:
Exception- if no parameters exist (yet)
-
toXML
public StringBuffer toXML()
Description copied from interface:StorableThis method returns an XML representation asStringBufferof an instance of the implementing class.
-
getLogPriorTerm
public double getLogPriorTerm()
Description copied from interface:DifferentiableStatisticalModelThis method computes a value that is proportional to
whereDifferentiableStatisticalModel.getESS()*DifferentiableStatisticalModel.getLogNormalizationConstant()+ Math.log( prior )prioris the prior for the parameters of this model.- Specified by:
getLogPriorTermin interfaceDifferentiableStatisticalModel- Specified by:
getLogPriorTermin interfaceStatisticalModel- Returns:
- a value that is proportional to
DifferentiableStatisticalModel.getESS()*DifferentiableStatisticalModel.getLogNormalizationConstant()+ Math.log( prior ). - See Also:
DifferentiableStatisticalModel.getESS(),DifferentiableStatisticalModel.getLogNormalizationConstant()
-
addGradientOfLogPriorTerm
public void addGradientOfLogPriorTerm(double[] grad, int start)Description copied from interface:DifferentiableStatisticalModelThis method computes the gradient ofDifferentiableStatisticalModel.getLogPriorTerm()for each parameter of this model. The results are added to the arraygradbeginning at indexstart.- Specified by:
addGradientOfLogPriorTermin interfaceDifferentiableStatisticalModel- Parameters:
grad- the array of gradientsstart- the start index in thegradarray, where the partial derivations for the parameters of this models shall be entered- See Also:
DifferentiableStatisticalModel.getLogPriorTerm()
-
getESS
public double getESS()
Description copied from interface:DifferentiableStatisticalModelReturns the equivalent sample size (ess) of this model, i.e. the equivalent sample size for the class or component that is represented by this model.- Specified by:
getESSin interfaceDifferentiableStatisticalModel- Returns:
- the equivalent sample size.
-
getPositionForParameter
public int getPositionForParameter(int index)
Returns the position in the sequence the parameterindexis responsible for.- Parameters:
index- the index of the parameter- Returns:
- the position in the sequence
-
getPositionDependentKMerProb
public double[] getPositionDependentKMerProb(Sequence kmer) throws Exception
Returns the probability ofkmerfor all possible positions in thisBayesianNetworkDiffSMstarting at positionkmer.getLength()-1.- Parameters:
kmer- the k-mer- Returns:
- the position-dependent probabilities of this k-mer for position
kmer.getLength()-1toAbstractDifferentiableSequenceScore.getLength()-1 - Throws:
Exception- if the method is called for non-Markov model structures
-
getSizeOfEventSpaceForRandomVariablesOfParameter
public int getSizeOfEventSpaceForRandomVariablesOfParameter(int index)
Description copied from interface:DifferentiableStatisticalModelReturns the size of the event space of the random variables that are affected by parameter no.index, i.e. the product of the sizes of the alphabets at the position of each random variable affected by parameterindex. For DNA alphabets this corresponds to 4 for a PWM, 16 for a WAM except position 0, ...- Specified by:
getSizeOfEventSpaceForRandomVariablesOfParameterin interfaceDifferentiableStatisticalModel- Parameters:
index- the index of the parameter- Returns:
- the size of the event space
-
initializeFunctionRandomly
public void initializeFunctionRandomly(boolean freeParams) throws ExceptionDescription copied from interface:DifferentiableSequenceScoreThis method initializes theDifferentiableSequenceScorerandomly. It has to create the underlying structure of theDifferentiableSequenceScore.- Specified by:
initializeFunctionRandomlyin interfaceDifferentiableSequenceScore- Parameters:
freeParams- indicates whether the (reduced) parameterization is used- Throws:
Exception- if something went wrong
-
isInitialized
public boolean isInitialized()
Description copied from interface:SequenceScoreThis method can be used to determine whether the instance is initialized. If the instance is initialized you should be able to invokeSequenceScore.getLogScoreFor(Sequence).- Specified by:
isInitializedin interfaceSequenceScore- Returns:
trueif the instance is initialized,falseotherwise
-
getPWM
public double[][] getPWM() throws ExceptionIf thisBayesianNetworkDiffSMis a PWM, i.e.structureMeasure=newInhomogeneousMarkov(0)}}, this method returns the normalized PWM as adoublearray of dimensionAbstractDifferentiableSequenceScore.getLength()x size-of-alphabet.- Returns:
- the PWM as a two-dimensional array
- Throws:
Exception- if this method is called for aBayesianNetworkDiffSMthat is not a PWM
-
getMaximalMarkovOrder
public byte getMaximalMarkovOrder() throws UnsupportedOperationExceptionDescription copied from interface:StatisticalModelThis method returns the maximal used Markov order, if possible.- Specified by:
getMaximalMarkovOrderin interfaceStatisticalModel- Overrides:
getMaximalMarkovOrderin classAbstractDifferentiableStatisticalModel- Returns:
- maximal used Markov order
- Throws:
UnsupportedOperationException- if the model can't give a proper answer
-
getMaximumScore
public double getMaximumScore() throws ExceptionReturns the maximum score of thisBayesianNetworkDiffSMreturned for a admissible input sequence. Currently only implemented for position weight matrices.- Returns:
- the maximum score
- Throws:
Exception- if the model is of higher order than 0
-
getCurrentParameterSet
public InstanceParameterSet getCurrentParameterSet() throws Exception
Description copied from interface:InstantiableFromParameterSetReturns theInstanceParameterSetthat has been used to instantiate the current instance of the implementing class. If the current instance was not created using anInstanceParameterSet, an equivalentInstanceParameterSetshould be returned, so that an instance created using thisInstanceParameterSetwould be in principle equal to the current instance.- Specified by:
getCurrentParameterSetin interfaceInstantiableFromParameterSet- Returns:
- the current
InstanceParameterSet - Throws:
Exception- if theInstanceParameterSetcould not be returned
-
emitDataSet
public DataSet emitDataSet(int numberOfSequences, int... seqLength) throws NotTrainedException, Exception
Description copied from interface:StatisticalModelThis method returns aDataSetobject containing artificial sequence(s).
There are two different possibilities to create a data set for a model with length 0 (homogeneous models).-
emitDataSet( int n, int l )should return a data set withnsequences of lengthl. -
emitDataSet( int n, int[] l )should return a data set withnsequences which have a sequence length corresponding to the entry in the given arrayl.
There are two different possibilities to create a data set for a model with length greater than 0 (inhomogeneous models).
emitDataSet( int n )andemitDataSet( int n, null )should return a data set withnsequences of length of the model (SequenceScore.getLength()).
The standard implementation throws anException.- Specified by:
emitDataSetin interfaceStatisticalModel- Overrides:
emitDataSetin classAbstractDifferentiableStatisticalModel- Parameters:
numberOfSequences- the number of sequences that should be contained in the returned data setseqLength- the length of the sequences for a homogeneous model; for an inhomogeneous model this parameter should benullor an array of size 0.- Returns:
- a
DataSetcontaining the artificial sequence(s) - Throws:
NotTrainedException- if the model is not trained yetException- if the emission did not succeed- See Also:
DataSet
-
-
toHtml
public String toHtml(NumberFormat nf)
Returns an HTML representation of thisBayesianNetworkDiffSM.- Parameters:
nf- the number format- Returns:
- the HTML representation
-
fillInfixScore
public void fillInfixScore(int[] seq, int start, int length, double[] scores)Description copied from interface:QuickScanningSequenceScoreComputes the position-wise scores of an infix of the sequenceseq(which must be encoded by the same alphabet as thisQuickScanningSequenceScore) beginning atstartand extending forlengthpositions. The scores are computed per position and filled into the provided arrayscores, which is of the same length asseq. This must be implemented such thatToolBox.sum(double...)applied toscorescomputed from0toseq.lengthreturns the same value asSequenceScore.getLogScoreFor(de.jstacs.data.sequences.Sequence)called on theIntSequencecreated fromseq.- Specified by:
fillInfixScorein interfaceQuickScanningSequenceScore- Parameters:
seq- the sequencestart- the start of the infixlength- the length of the infixscores- the array of scores to be (partly) filled
-
-
![\[\log \frac{\partial Z(\underline{\lambda})}{\partial \lambda_{parameterindex}}\]](images/DifferentiableStatisticalModel_LaTeXilb10_1.png)