 de.jstacs.scoringFunctions.AbstractNormalizableScoringFunction
de.jstacs.scoringFunctions.AbstractNormalizableScoringFunction
 de.jstacs.scoringFunctions.IndependentProductScoringFunction
de.jstacs.scoringFunctions.IndependentProductScoringFunction
|
||||||||||
| PREV CLASS NEXT CLASS | FRAMES NO FRAMES | |||||||||
| SUMMARY: NESTED | FIELD | CONSTR | METHOD | DETAIL: FIELD | CONSTR | METHOD | |||||||||
java.lang.Objectde.jstacs.scoringFunctions.AbstractNormalizableScoringFunction
de.jstacs.scoringFunctions.IndependentProductScoringFunction
public class IndependentProductScoringFunction
This class enables the user to model parts of a sequence independent of each
other. For instance, the first part of the sequence is modeled by the first
NormalizableScoringFunction and has the length of the first
NormalizableScoringFunction, the second part starts directly after
the first part, is modeled by the second NormalizableScoringFunction
... etc. It is also possible to use a NormalizableScoringFunction for
more than one sequence part and in both orientations (if possible).
It is important to set the equivalent sample size (ESS) of each instance carefully, i.e., corresponding to the ESS of the parts.
| Nested Class Summary |
|---|
| Nested classes/interfaces inherited from interface de.jstacs.motifDiscovery.MotifDiscoverer |
|---|
MotifDiscoverer.KindOfProfile |
| Field Summary |
|---|
| Fields inherited from class de.jstacs.scoringFunctions.AbstractNormalizableScoringFunction |
|---|
alphabets, length, r |
| Fields inherited from interface de.jstacs.scoringFunctions.ScoringFunction |
|---|
UNKNOWN |
| Constructor Summary | |
|---|---|
IndependentProductScoringFunction(double ess,
boolean plugIn,
NormalizableScoringFunction... functions)
This constructor creates an instance of an IndependentProductScoringFunction from a given series of
independent NormalizableScoringFunctions. |
|
IndependentProductScoringFunction(double ess,
boolean plugIn,
NormalizableScoringFunction[] functions,
int[] length)
This constructor creates an instance of an IndependentProductScoringFunction from given series of
independent NormalizableScoringFunctions and lengths. |
|
IndependentProductScoringFunction(double ess,
boolean plugIn,
NormalizableScoringFunction[] functions,
int[] index,
int[] length,
boolean[] reverse)
This is the main constructor. |
|
IndependentProductScoringFunction(StringBuffer source)
This is the constructor for the interface Storable. |
|
| Method Summary | |
|---|---|
void |
addGradientOfLogPriorTerm(double[] grad,
int start)
This method computes the gradient of NormalizableScoringFunction.getLogPriorTerm() for each
parameter of this model. |
void |
adjustHiddenParameters(int index,
Sample[] data,
double[][] weights)
Adjusts all hidden parameters including duration and mixture parameters according to the current values of the remaining parameters. |
IndependentProductScoringFunction |
clone()
Creates a clone (deep copy) of the current ScoringFunction
instance. |
int |
extractSequenceParts(int scoringFunctionIndex,
Sample[] data,
Sample[] result)
This method extracts the corresponding Sequence parts for a specific ScoringFunction. |
double[][] |
extractWeights(int number,
double[][] weights)
This method creates the weights for extractSequenceParts(int, Sample[], Sample[]). |
protected void |
fromXML(StringBuffer rep)
This method is called in the constructor for the Storable
interface to create a scoring function from a StringBuffer. |
double[] |
getCurrentParameterValues()
Returns a double array of dimension
ScoringFunction.getNumberOfParameters() containing the current parameter values. |
double |
getEss()
Returns the equivalent sample size (ess) of this model, i.e. the equivalent sample size for the class or component that is represented by this model. |
NormalizableScoringFunction[] |
getFunctions()
This method returns a deep copy of the internally used NormalizableScoringFunction. |
int |
getGlobalIndexOfMotifInComponent(int component,
int motif)
Returns the global index of the motif used in
component. |
int |
getIndexOfMaximalComponentFor(Sequence sequence)
Returns the index of the component with the maximal score for the sequence sequence. |
int[] |
getIndices()
This method returns a deep copy of the internally used indices of the NormalizableScoringFunction for the parts. |
String |
getInstanceName()
Returns a short instance name. |
double |
getLogNormalizationConstant()
Returns the logarithm of the sum of the scores over all sequences of the event space. |
double |
getLogPartialNormalizationConstant(int parameterIndex)
Returns the logarithm of the partial normalization constant for the parameter with index parameterIndex. |
double |
getLogPriorTerm()
This method computes a value that is proportional to
where prior is the prior for the parameters of this model. |
double |
getLogScore(Sequence seq,
int start)
Returns the logarithmic score for the Sequence seq
beginning at position start in the Sequence. |
double |
getLogScoreAndPartialDerivation(Sequence seq,
int start,
IntList indices,
DoubleList partialDer)
Returns the logarithmic score for a Sequence beginning at
position start in the Sequence and fills lists with
the indices and the partial derivations. |
int |
getMotifLength(int motif)
This method returns the length of the motif with index motif
. |
int |
getNumberOfComponents()
Returns the number of components in this MotifDiscoverer. |
int |
getNumberOfMotifs()
Returns the number of motifs for this MotifDiscoverer. |
int |
getNumberOfMotifsInComponent(int component)
Returns the number of motifs that are used in the component component of this MotifDiscoverer. |
int |
getNumberOfParameters()
Returns the number of parameters in this ScoringFunction. |
int |
getNumberOfRecommendedStarts()
This method returns the number of recommended optimization starts. |
int[] |
getPartialLengths()
This method returns a deep copy of the internally used partial lengths of the parts. |
double[] |
getProfileOfScoresFor(int component,
int motif,
Sequence sequence,
int startpos,
MotifDiscoverer.KindOfProfile dist)
Returns the profile of the scores for component component
and motif motif at all possible start positions of the motif
in the sequence sequence beginning at startpos. |
boolean[] |
getReverseSwitches()
This method returns a deep copy of the internally used switches for the parts whether to use the corresponding NormalizableScoringFunction forward or as reverse complement. |
int |
getSizeOfEventSpaceForRandomVariablesOfParameter(int index)
Returns the size of the event space of the random variables that are affected by parameter no. |
double[] |
getStrandProbabilitiesFor(int component,
int motif,
Sequence sequence,
int startpos)
This method returns the probabilities of the strand orientations for a given subsequence if it is considered as site of the motif model in a specific component. |
void |
initializeFunction(int index,
boolean freeParams,
Sample[] data,
double[][] weights)
This method creates the underlying structure of the ScoringFunction. |
void |
initializeFunctionRandomly(boolean freeParams)
This method initializes the ScoringFunction randomly. |
void |
initializeMotif(int motifIndex,
Sample data,
double[] weights)
This method allows to initialize the model of a motif manually using a weighted sample. |
void |
initializeMotifRandomly(int motif)
This method initializes the motif with index motif randomly using for instance ScoringFunction.initializeFunctionRandomly(boolean). |
boolean |
isInitialized()
This method can be used to determine whether the model is initialized. |
boolean |
isNormalized()
This method indicates whether the implemented score is already normalized to 1 or not. |
boolean |
modifyMotif(int motifIndex,
int offsetLeft,
int offsetRight)
Manually modifies the motif model with index motifIndex. |
void |
setParameters(double[] params,
int start)
This method sets the internal parameters to the values of params between start and
start + |
String |
toString()
|
StringBuffer |
toXML()
This method returns an XML representation as StringBuffer of an
instance of the implementing class. |
| Methods inherited from class de.jstacs.scoringFunctions.AbstractNormalizableScoringFunction |
|---|
getAlphabetContainer, getInitialClassParam, getLength, getLogScore, getLogScoreAndPartialDerivation, getNumberOfStarts, isNormalized |
| Methods inherited from class java.lang.Object |
|---|
equals, finalize, getClass, hashCode, notify, notifyAll, wait, wait, wait |
| Constructor Detail |
|---|
public IndependentProductScoringFunction(double ess,
boolean plugIn,
NormalizableScoringFunction... functions)
throws CloneNotSupportedException,
IllegalArgumentException,
WrongAlphabetException
IndependentProductScoringFunction from a given series of
independent NormalizableScoringFunctions. The length that is
modeled by each component is determined by
ScoringFunction.getLength(). So the length should not be 0.
ess - the equivalent sample sizeplugIn - whether to use plugIn parameters for the parts, otherwise the last parameters are used for parts that are instance of HomogeneousScoringFunctionfunctions - the components, i.e. the given series of independent
NormalizableScoringFunctions
CloneNotSupportedException - if at least one element of functions could not
be cloned
IllegalArgumentException - if at least one component has length 0 or if the
equivalent sample size (ess) is smaller than zero (0)
WrongAlphabetException - if the user tries to use an alphabet for a reverse complement that can not be used for a reverse complement.IndependentProductScoringFunction(double, boolean, NormalizableScoringFunction[], int[])
public IndependentProductScoringFunction(double ess,
boolean plugIn,
NormalizableScoringFunction[] functions,
int[] length)
throws CloneNotSupportedException,
IllegalArgumentException,
WrongAlphabetException
IndependentProductScoringFunction from given series of
independent NormalizableScoringFunctions and lengths.
ess - the equivalent sample sizeplugIn - whether to use plugIn parameters for the parts, otherwise the last parameters are used for parts that are instance of HomogeneousScoringFunctionfunctions - the components, i.e. the given series of independent
NormalizableScoringFunctionslength - the lengths, one for each component
CloneNotSupportedException - if at least one component could not be cloned
IllegalArgumentException - if the lengths and the components are not matching or if the
equivalent sample size (ess) is smaller than zero (0)
WrongAlphabetException - if the user tries to use an alphabet for a reverse complement that can not be used for a reverse complement.IndependentProductScoringFunction(double, boolean, NormalizableScoringFunction[], int[], int[], boolean[])
public IndependentProductScoringFunction(double ess,
boolean plugIn,
NormalizableScoringFunction[] functions,
int[] index,
int[] length,
boolean[] reverse)
throws CloneNotSupportedException,
IllegalArgumentException,
WrongAlphabetException
ess - the equivalent sample sizeplugIn - whether to use plugIn parameters for the parts, otherwise the last parameters are used for parts that are instance of HomogeneousScoringFunctionfunctions - the NormalizableScoringFunctionindex - the index of the NormalizableScoringFunction at each partlength - the length of each partreverse - a switch whether to use it directly or the reverse complementary strand
CloneNotSupportedException - if at least one component could not be cloned
IllegalArgumentException - if the lengths and the components are not matching or if the
equivalent sample size (ess) is smaller than zero (0)
WrongAlphabetException - if the user tries to use an alphabet for a reverse complement that can not be used for a reverse complement.
public IndependentProductScoringFunction(StringBuffer source)
throws NonParsableException
Storable.
Creates a new IndependentProductScoringFunction out of a
StringBuffer as returned by toXML().
source - the XML representation as StringBuffer
NonParsableException - if the XML representation could not be parsed| Method Detail |
|---|
public IndependentProductScoringFunction clone()
throws CloneNotSupportedException
ScoringFunctionScoringFunction
instance.
clone in interface MotifDiscovererclone in interface ScoringFunctionclone in class AbstractNormalizableScoringFunctionScoringFunction
CloneNotSupportedException - if something went wrong while cloning the
ScoringFunctionCloneablepublic int getSizeOfEventSpaceForRandomVariablesOfParameter(int index)
NormalizableScoringFunctionindex, i.e. the product of the
sizes of the alphabets at the position of each random variable affected
by parameter index. For DNA alphabets this corresponds to 4
for a PWM, 16 for a WAM except position 0, ...
getSizeOfEventSpaceForRandomVariablesOfParameter in interface NormalizableScoringFunctionindex - the index of the parameter
public double getLogNormalizationConstant()
NormalizableScoringFunction
getLogNormalizationConstant in interface NormalizableScoringFunction
public double getLogPartialNormalizationConstant(int parameterIndex)
throws Exception
NormalizableScoringFunctionparameterIndex. This is the logarithm of the partial derivation of the
normalization constant for the parameter with index
parameterIndex,
![\[\log \frac{\partial Z(\underline{\lambda})}{\partial \lambda_{parameterindex}}\]](images/NormalizableScoringFunction_LaTeXilb8_1.png)
getLogPartialNormalizationConstant in interface NormalizableScoringFunctionparameterIndex - the index of the parameter
Exception - if something went wrong with the normalizationNormalizableScoringFunction.getLogNormalizationConstant()public double getEss()
NormalizableScoringFunction
getEss in interface NormalizableScoringFunction
public void initializeFunction(int index,
boolean freeParams,
Sample[] data,
double[][] weights)
throws Exception
ScoringFunctionScoringFunction.
initializeFunction in interface ScoringFunctionindex - the index of the class the ScoringFunction modelsfreeParams - indicates whether the (reduced) parameterization is useddata - the samplesweights - the weights of the sequences in the samples
Exception - if something went wrong
public int extractSequenceParts(int scoringFunctionIndex,
Sample[] data,
Sample[] result)
throws Exception
Sequence parts for a specific ScoringFunction.
scoringFunctionIndex - the index of the ScoringFunctiondata - the original dataresult - an array for the resulting Samples of Sequences; has to have same length as data
ScoringFunction was used
Exception - if the Sample can not be created
public double[][] extractWeights(int number,
double[][] weights)
extractSequenceParts(int, Sample[], Sample[]).
number - the number how often the weights should be copied after each other.weights - the original weights
null)extractSequenceParts(int, Sample[], Sample[])
protected void fromXML(StringBuffer rep)
throws NonParsableException
AbstractNormalizableScoringFunctionStorable
interface to create a scoring function from a StringBuffer.
fromXML in class AbstractNormalizableScoringFunctionrep - the XML representation as StringBuffer
NonParsableException - if the StringBuffer could not be parsedAbstractNormalizableScoringFunction.AbstractNormalizableScoringFunction(StringBuffer)public String getInstanceName()
ScoringFunction
getInstanceName in interface ScoringFunction
public NormalizableScoringFunction[] getFunctions()
throws Exception
NormalizableScoringFunction.
NormalizableScoringFunction
Exception - if at least one NormalizableScoringFunction could not be clonedgetIndices(),
getPartialLengths(),
getReverseSwitches()public int[] getIndices()
NormalizableScoringFunction for the parts.
NormalizableScoringFunction for the partsgetFunctions(),
getPartialLengths(),
getReverseSwitches()public int[] getPartialLengths()
getFunctions(),
getIndices(),
getReverseSwitches()public boolean[] getReverseSwitches()
NormalizableScoringFunction forward or as reverse complement.
NormalizableScoringFunction forward or as reverse complementgetFunctions(),
getIndices(),
getPartialLengths()
public double[] getCurrentParameterValues()
throws Exception
ScoringFunctiondouble array of dimension
ScoringFunction.getNumberOfParameters() containing the current parameter values.
If one likes to use these parameters to start an optimization it is
highly recommended to invoke
ScoringFunction.initializeFunction(int, boolean, Sample[], double[][]) before.
After an optimization this method can be used to get the current
parameter values.
getCurrentParameterValues in interface ScoringFunctionException - if no parameters exist (yet)
public double getLogScore(Sequence seq,
int start)
ScoringFunctionSequence seq
beginning at position start in the Sequence.
getLogScore in interface ScoringFunctionseq - the Sequencestart - the start position in the Sequence
Sequence
public double getLogScoreAndPartialDerivation(Sequence seq,
int start,
IntList indices,
DoubleList partialDer)
ScoringFunctionSequence beginning at
position start in the Sequence and fills lists with
the indices and the partial derivations.
getLogScoreAndPartialDerivation in interface ScoringFunctionseq - the Sequencestart - the start position in the Sequenceindices - an IntList of indices, after method invocation the
list should contain the indices i where
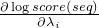 is not zero
is not zeropartialDer - a DoubleList of partial derivations, after method
invocation the list should contain the corresponding
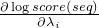 that are not zero
that are not zero
Sequencepublic int getNumberOfParameters()
ScoringFunctionScoringFunction. If the
number of parameters is not known yet, the method returns
ScoringFunction.UNKNOWN.
getNumberOfParameters in interface ScoringFunctionScoringFunctionScoringFunction.UNKNOWNpublic int getNumberOfRecommendedStarts()
ScoringFunction
getNumberOfRecommendedStarts in interface ScoringFunctiongetNumberOfRecommendedStarts in class AbstractNormalizableScoringFunction
public void setParameters(double[] params,
int start)
ScoringFunctionparams between start and
start + ScoringFunction.getNumberOfParameters() - 1
setParameters in interface ScoringFunctionparams - the new parametersstart - the start index in paramspublic StringBuffer toXML()
StorableStringBuffer of an
instance of the implementing class.
toXML in interface Storablepublic String toString()
toString in class Objectpublic double getLogPriorTerm()
NormalizableScoringFunction
NormalizableScoringFunction.getEss() * NormalizableScoringFunction.getLogNormalizationConstant() + Math.log( prior )
prior is the prior for the parameters of this model.
getLogPriorTerm in interface NormalizableScoringFunctionNormalizableScoringFunction.getEss() * NormalizableScoringFunction.getLogNormalizationConstant() + Math.log( prior ).NormalizableScoringFunction.getEss(),
NormalizableScoringFunction.getLogNormalizationConstant()
public void addGradientOfLogPriorTerm(double[] grad,
int start)
throws Exception
NormalizableScoringFunctionNormalizableScoringFunction.getLogPriorTerm() for each
parameter of this model. The results are added to the array
grad beginning at index start.
addGradientOfLogPriorTerm in interface NormalizableScoringFunctiongrad - the array of gradientsstart - the start index in the grad array, where the
partial derivations for the parameters of this models shall be
entered
Exception - if something went wrong with the computing of the gradientsNormalizableScoringFunction.getLogPriorTerm()public boolean isInitialized()
ScoringFunctionScoringFunction.initializeFunction(int, boolean, Sample[], double[][]).
isInitialized in interface ScoringFunctiontrue if the model is initialized, false
otherwise
public void initializeFunctionRandomly(boolean freeParams)
throws Exception
ScoringFunctionScoringFunction randomly. It has to
create the underlying structure of the ScoringFunction.
initializeFunctionRandomly in interface ScoringFunctionfreeParams - indicates whether the (reduced) parameterization is used
Exception - if something went wrong
public void initializeMotif(int motifIndex,
Sample data,
double[] weights)
throws Exception
MutableMotifDiscoverer
initializeMotif in interface MutableMotifDiscoverermotifIndex - the index of the motif in the motif discovererdata - the sample of sequencesweights - either null or an array of length data.getNumberofElements() with non-negative weights.
Exception - if initialize was not possible
public void initializeMotifRandomly(int motif)
throws Exception
MutableMotifDiscoverermotif randomly using for instance ScoringFunction.initializeFunctionRandomly(boolean).
Furthermore, if available, it also initializes the positional distribution.
initializeMotifRandomly in interface MutableMotifDiscoverermotif - the index of the motif
Exception - either if the index is wrong or if it is thrown by the method ScoringFunction.initializeFunctionRandomly(boolean)
public boolean modifyMotif(int motifIndex,
int offsetLeft,
int offsetRight)
throws Exception
MutableMotifDiscoverermotifIndex. The two offsets offsetLeft and offsetRight
define how many positions the left or right border positions shall be moved. Negative numbers indicate moves to the left while positive
numbers correspond to moves to the right. The distribution for sequences to the left and right side of the motif shall be computed internally.
modifyMotif in interface MutableMotifDiscoverermotifIndex - the index of the motif in the motif discovereroffsetLeft - the offset on the left sideoffsetRight - the offset on the right side
true if the motif model was modified otherwise false
Exception - if some unexpected error occurred during the modificationMutableMotifDiscoverer.modifyMotif(int, int, int),
Mutable.modify(int, int)
public int getGlobalIndexOfMotifInComponent(int component,
int motif)
MotifDiscoverermotif used in
component. The index returned must be at least 0 and less
than MotifDiscoverer.getNumberOfMotifs().
getGlobalIndexOfMotifInComponent in interface MotifDiscoverercomponent - the component indexmotif - the motif index in the component
motif in component
public int getIndexOfMaximalComponentFor(Sequence sequence)
throws Exception
MotifDiscoverersequence.
getIndexOfMaximalComponentFor in interface MotifDiscoverersequence - the given sequence
Exception - if the index could not be computed for any reasonspublic int getMotifLength(int motif)
MotifDiscoverermotif
.
getMotifLength in interface MotifDiscoverermotif - the index of the motif
motifpublic int getNumberOfComponents()
MotifDiscovererMotifDiscoverer.
getNumberOfComponents in interface MotifDiscovererpublic int getNumberOfMotifs()
MotifDiscovererMotifDiscoverer.
getNumberOfMotifs in interface MotifDiscovererpublic int getNumberOfMotifsInComponent(int component)
MotifDiscoverercomponent of this MotifDiscoverer.
getNumberOfMotifsInComponent in interface MotifDiscoverercomponent - the component of the MotifDiscoverer
public double[] getProfileOfScoresFor(int component,
int motif,
Sequence sequence,
int startpos,
MotifDiscoverer.KindOfProfile dist)
throws Exception
MotifDiscoverercomponent
and motif motif at all possible start positions of the motif
in the sequence sequence beginning at startpos.
This array should be of length sequence.length() - startpos - motifs[motif].length() + 1.
getProfileOfScoresFor in interface MotifDiscoverercomponent - the component indexmotif - the index of the motif in the componentsequence - the given sequencestartpos - the start position in the sequencedist - indicates the kind of profile
Exception - if the score could not be computed for any reasons
public double[] getStrandProbabilitiesFor(int component,
int motif,
Sequence sequence,
int startpos)
throws Exception
MotifDiscoverer
getStrandProbabilitiesFor in interface MotifDiscoverercomponent - the component indexmotif - the index of the motif in the componentsequence - the given sequencestartpos - the start position in the sequence
Exception - if the strand could not be computed for any reasonspublic boolean isNormalized()
NormalizableScoringFunctionfalse.
isNormalized in interface NormalizableScoringFunctionisNormalized in class AbstractNormalizableScoringFunctiontrue if the implemented score is already normalized
to 1, false otherwise
public void adjustHiddenParameters(int index,
Sample[] data,
double[][] weights)
throws Exception
MutableMotifDiscoverer
adjustHiddenParameters in interface MutableMotifDiscovererindex - the index of the class of this MutableMotifDiscovererdata - the array of data for all classesweights - the weights for all sequences in data
Exception - thrown if the hidden parameters could not be adjusted
|
||||||||||
| PREV CLASS NEXT CLASS | FRAMES NO FRAMES | |||||||||
| SUMMARY: NESTED | FIELD | CONSTR | METHOD | DETAIL: FIELD | CONSTR | METHOD | |||||||||