 de.jstacs.scoringFunctions.AbstractNormalizableScoringFunction
de.jstacs.scoringFunctions.AbstractNormalizableScoringFunction
 de.jstacs.scoringFunctions.AbstractVariableLengthScoringFunction
de.jstacs.scoringFunctions.AbstractVariableLengthScoringFunction
 de.jstacs.scoringFunctions.CMMScoringFunction
de.jstacs.scoringFunctions.CMMScoringFunction
|
||||||||||
| PREV CLASS NEXT CLASS | FRAMES NO FRAMES | |||||||||
| SUMMARY: NESTED | FIELD | CONSTR | METHOD | DETAIL: FIELD | CONSTR | METHOD | |||||||||
java.lang.Objectde.jstacs.scoringFunctions.AbstractNormalizableScoringFunction
de.jstacs.scoringFunctions.AbstractVariableLengthScoringFunction
de.jstacs.scoringFunctions.CMMScoringFunction
public class CMMScoringFunction
This scoring function implements a cyclic Markov model of arbitrary order and periodicity for any sequence length. The scoring function uses the parametrization of Meila.
| Field Summary |
|---|
| Fields inherited from class de.jstacs.scoringFunctions.AbstractNormalizableScoringFunction |
|---|
alphabets, length, r |
| Fields inherited from interface de.jstacs.scoringFunctions.ScoringFunction |
|---|
UNKNOWN |
| Constructor Summary | |
|---|---|
CMMScoringFunction(AlphabetContainer alphabets,
double[] frameHyper,
double[][][] hyper,
boolean plugIn,
boolean optimize,
int starts,
int initFrame)
This constructor allows to create an instance with specific hyper-parameters for all conditional distributions. |
|
CMMScoringFunction(AlphabetContainer alphabets,
int order,
int period,
double classEss,
double[] sumOfHyperParams,
boolean plugIn,
boolean optimize,
int starts,
int initFrame)
The main constructor. |
|
CMMScoringFunction(StringBuffer source)
This is the constructor for Storable. |
|
| Method Summary | |
|---|---|
void |
addGradientOfLogPriorTerm(double[] grad,
int start)
This method computes the gradient of NormalizableScoringFunction.getLogPriorTerm() for each
parameter of this model. |
CMMScoringFunction |
clone()
Creates a clone (deep copy) of the current ScoringFunction
instance. |
protected void |
fromXML(StringBuffer xml)
This method is called in the constructor for the Storable
interface to create a scoring function from a StringBuffer. |
double[] |
getCurrentParameterValues()
Returns a double array of dimension
ScoringFunction.getNumberOfParameters() containing the current parameter values. |
double |
getEss()
Returns the equivalent sample size (ess) of this model, i.e. the equivalent sample size for the class or component that is represented by this model. |
static double[][][] |
getHyperParams(int alphabetSize,
int length,
double ess,
double[] frameProb,
double[][][] prob)
This method returns the hyper-parameters for a model given some a-priori probabilities. |
String |
getInstanceName()
Returns a short instance name. |
double |
getLogNormalizationConstant(int length)
This method returns the logarithm of the normalization constant for a given sequence length. |
double |
getLogPartialNormalizationConstant(int parameterIndex,
int length)
This method returns the logarithm of the partial normalization constant for a given parameter index and a sequence length. |
double |
getLogPriorTerm()
This method computes a value that is proportional to
where prior is the prior for the parameters of this model. |
double |
getLogScore(Sequence seq,
int start,
int length)
This method computes the logarithm of the score for a given subsequence. |
double |
getLogScoreAndPartialDerivation(Sequence seq,
int start,
int length,
IntList indices,
DoubleList dList)
This method computes the logarithm of the score and the partial derivations for a given subsequence. |
int |
getNumberOfParameters()
Returns the number of parameters in this ScoringFunction. |
int |
getNumberOfRecommendedStarts()
This method returns the number of recommended optimization starts. |
int |
getSizeOfEventSpaceForRandomVariablesOfParameter(int index)
Returns the size of the event space of the random variables that are affected by parameter no. |
void |
initializeFunction(int index,
boolean freeParams,
Sample[] data,
double[][] weights)
This method creates the underlying structure of the ScoringFunction. |
void |
initializeFunctionRandomly(boolean freeParams)
This method initializes the ScoringFunction randomly. |
boolean |
isInitialized()
This method can be used to determine whether the model is initialized. |
boolean |
isNormalized()
This method indicates whether the implemented score is already normalized to 1 or not. |
void |
setFrameParameterOptimization(boolean optimize)
This method enables the user to choose whether the frame parameters should be optimized or not. |
void |
setParameterOptimization(boolean optimize)
This method enables the user to choose whether the parameters should be optimized or not. |
void |
setParameters(double[] params,
int start)
This method sets the internal parameters to the values of params between start and
start + |
void |
setStatisticForHyperparameters(int[] length,
double[] weight)
This method sets the hyperparameters for the model parameters by evaluating the given statistic. |
String |
toString()
|
StringBuffer |
toXML()
This method returns an XML representation as StringBuffer of an
instance of the implementing class. |
| Methods inherited from class de.jstacs.scoringFunctions.AbstractVariableLengthScoringFunction |
|---|
getLogNormalizationConstant, getLogPartialNormalizationConstant, getLogScore, getLogScoreAndPartialDerivation |
| Methods inherited from class de.jstacs.scoringFunctions.AbstractNormalizableScoringFunction |
|---|
getAlphabetContainer, getInitialClassParam, getLength, getLogScore, getLogScoreAndPartialDerivation, getNumberOfStarts, isNormalized |
| Methods inherited from class java.lang.Object |
|---|
equals, finalize, getClass, hashCode, notify, notifyAll, wait, wait, wait |
| Methods inherited from interface de.jstacs.scoringFunctions.NormalizableScoringFunction |
|---|
getInitialClassParam |
| Methods inherited from interface de.jstacs.scoringFunctions.ScoringFunction |
|---|
getAlphabetContainer, getLength, getLogScore, getLogScoreAndPartialDerivation |
| Constructor Detail |
|---|
public CMMScoringFunction(AlphabetContainer alphabets,
int order,
int period,
double classEss,
double[] sumOfHyperParams,
boolean plugIn,
boolean optimize,
int starts,
int initFrame)
alphabets - the alphabet containerorder - the oder of the model (has to be non-negative)period - the periodclassEss - the ess of the classsumOfHyperParams - the sum of the hyper parameter for each order (length has to be order+1, each entry has to be non-negative), the sum also sums over the periodplugIn - a switch which enables to used the MAP-parameters as plug-in parametersoptimize - a switch which enables to optimize or fix the parametersstarts - the number of recommended startsinitFrame - the frame which should be used for plug-in initialization, negative for random initializationgetHyperParams(int, int, double, double[], double[][][]),
CMMScoringFunction(AlphabetContainer, double[], double[][][], boolean, boolean, int, int)
public CMMScoringFunction(AlphabetContainer alphabets,
double[] frameHyper,
double[][][] hyper,
boolean plugIn,
boolean optimize,
int starts,
int initFrame)
alphabets - the alphabet containerframeHyper - the hyper-parameters for the frame, the length of this array also defines the period of the modelhyper - the hyper-parameters for each frameplugIn - a switch which enables to used the MAP-parameters as plug-in parametersoptimize - a switch which enables to optimize or fix the parametersstarts - the number of recommended startsinitFrame - the frame which should be used for plug-in initialization, negative for random initialization
public CMMScoringFunction(StringBuffer source)
throws NonParsableException
Storable.
source - the xml representation
NonParsableException - if the representation could not be parsed.| Method Detail |
|---|
public static double[][][] getHyperParams(int alphabetSize,
int length,
double ess,
double[] frameProb,
double[][][] prob)
alphabetSize - the size of the alphabetlength - the expected sequence lengthess - the equivalent sample size (ess) of the modelframeProb - the a-priori probabilities for each frameprob - the a-priori probabilities for each frame and order
public CMMScoringFunction clone()
throws CloneNotSupportedException
ScoringFunctionScoringFunction
instance.
clone in interface ScoringFunctionclone in class AbstractNormalizableScoringFunctionScoringFunction
CloneNotSupportedException - if something went wrong while cloning the
ScoringFunctionpublic String getInstanceName()
ScoringFunction
public double getLogScore(Sequence seq,
int start,
int length)
VariableLengthScoringFunction
seq - the Sequencestart - the start index in the Sequencelength - the length of the Sequence beginning at start
ScoringFunction.getLogScore(Sequence,
int)
public double getLogScoreAndPartialDerivation(Sequence seq,
int start,
int length,
IntList indices,
DoubleList dList)
VariableLengthScoringFunction
seq - the Sequencestart - the start index in the Sequencelength - the end index in the Sequenceindices - an IntList of indices, after method invocation the
list should contain the indices i where
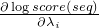 is not zero
is not zerodList - a DoubleList of partial derivations, after method
invocation the list should contain the corresponding
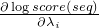 that are not zero
that are not zero
ScoringFunction.getLogScoreAndPartialDerivation(Sequence, int,
IntList, DoubleList)public int getNumberOfParameters()
ScoringFunctionScoringFunction. If the
number of parameters is not known yet, the method returns
ScoringFunction.UNKNOWN.
ScoringFunctionScoringFunction.UNKNOWN
public void setParameters(double[] params,
int start)
ScoringFunctionparams between start and
start + ScoringFunction.getNumberOfParameters() - 1
params - the new parametersstart - the start index in paramspublic StringBuffer toXML()
StorableStringBuffer of an
instance of the implementing class.
public double[] getCurrentParameterValues()
ScoringFunctiondouble array of dimension
ScoringFunction.getNumberOfParameters() containing the current parameter values.
If one likes to use these parameters to start an optimization it is
highly recommended to invoke
ScoringFunction.initializeFunction(int, boolean, Sample[], double[][]) before.
After an optimization this method can be used to get the current
parameter values.
public void initializeFunction(int index,
boolean freeParams,
Sample[] data,
double[][] weights)
ScoringFunctionScoringFunction.
index - the index of the class the ScoringFunction modelsfreeParams - indicates whether the (reduced) parameterization is useddata - the samplesweights - the weights of the sequences in the samplespublic void initializeFunctionRandomly(boolean freeParams)
ScoringFunctionScoringFunction randomly. It has to
create the underlying structure of the ScoringFunction.
freeParams - indicates whether the (reduced) parameterization is used
protected void fromXML(StringBuffer xml)
throws NonParsableException
AbstractNormalizableScoringFunctionStorable
interface to create a scoring function from a StringBuffer.
fromXML in class AbstractNormalizableScoringFunctionxml - the XML representation as StringBuffer
NonParsableException - if the StringBuffer could not be parsedAbstractNormalizableScoringFunction.AbstractNormalizableScoringFunction(StringBuffer)public int getSizeOfEventSpaceForRandomVariablesOfParameter(int index)
NormalizableScoringFunctionindex, i.e. the product of the
sizes of the alphabets at the position of each random variable affected
by parameter index. For DNA alphabets this corresponds to 4
for a PWM, 16 for a WAM except position 0, ...
index - the index of the parameter
public double getLogNormalizationConstant(int length)
VariableLengthScoringFunction
length - the sequence length
NormalizableScoringFunction.getLogNormalizationConstant()
public double getLogPartialNormalizationConstant(int parameterIndex,
int length)
throws Exception
VariableLengthScoringFunction
parameterIndex - the index of the parameterlength - the sequence length
Exception - if something went wrongNormalizableScoringFunction.getLogPartialNormalizationConstant(int)public double getEss()
NormalizableScoringFunction
public String toString()
toString in class Objectpublic double getLogPriorTerm()
NormalizableScoringFunction
NormalizableScoringFunction.getEss() * NormalizableScoringFunction.getLogNormalizationConstant() + Math.log( prior )
prior is the prior for the parameters of this model.
NormalizableScoringFunction.getEss() * NormalizableScoringFunction.getLogNormalizationConstant() + Math.log( prior ).NormalizableScoringFunction.getEss(),
NormalizableScoringFunction.getLogNormalizationConstant()
public void addGradientOfLogPriorTerm(double[] grad,
int start)
NormalizableScoringFunctionNormalizableScoringFunction.getLogPriorTerm() for each
parameter of this model. The results are added to the array
grad beginning at index start.
grad - the array of gradientsstart - the start index in the grad array, where the
partial derivations for the parameters of this models shall be
enteredNormalizableScoringFunction.getLogPriorTerm()public boolean isNormalized()
NormalizableScoringFunctionfalse.
isNormalized in interface NormalizableScoringFunctionisNormalized in class AbstractNormalizableScoringFunctiontrue if the implemented score is already normalized
to 1, false otherwisepublic boolean isInitialized()
ScoringFunctionScoringFunction.initializeFunction(int, boolean, Sample[], double[][]).
true if the model is initialized, false
otherwisepublic int getNumberOfRecommendedStarts()
ScoringFunction
getNumberOfRecommendedStarts in interface ScoringFunctiongetNumberOfRecommendedStarts in class AbstractNormalizableScoringFunctionpublic void setParameterOptimization(boolean optimize)
optimize - the switch for optimization of the parameterspublic void setFrameParameterOptimization(boolean optimize)
optimize - the switch for optimization of the frame parameters
public void setStatisticForHyperparameters(int[] length,
double[] weight)
throws Exception
VariableLengthScoringFunctionlength) and how often (
weight) they have been seen.
length - the non-negative lengths of the sequencesweight - the non-negative weight for the corresponding sequence
Exception - if something went wrongMutable
|
||||||||||
| PREV CLASS NEXT CLASS | FRAMES NO FRAMES | |||||||||
| SUMMARY: NESTED | FIELD | CONSTR | METHOD | DETAIL: FIELD | CONSTR | METHOD | |||||||||