 de.jstacs.classifiers.AbstractClassifier
de.jstacs.classifiers.AbstractClassifier
 de.jstacs.classifiers.AbstractScoreBasedClassifier
de.jstacs.classifiers.AbstractScoreBasedClassifier
 de.jstacs.classifiers.differentiableSequenceScoreBased.sampling.SamplingScoreBasedClassifier
de.jstacs.classifiers.differentiableSequenceScoreBased.sampling.SamplingScoreBasedClassifier
|
||||||||||
| PREV CLASS NEXT CLASS | FRAMES NO FRAMES | |||||||||
| SUMMARY: NESTED | FIELD | CONSTR | METHOD | DETAIL: FIELD | CONSTR | METHOD | |||||||||
java.lang.Objectde.jstacs.classifiers.AbstractClassifier
de.jstacs.classifiers.AbstractScoreBasedClassifier
de.jstacs.classifiers.differentiableSequenceScoreBased.sampling.SamplingScoreBasedClassifier
public abstract class SamplingScoreBasedClassifier
A classifier that samples the parameters of SamplingDifferentiableStatisticalModels by the Metropolis-Hastings algorithm.
The distribution the parameters are sampled from is the distribution 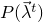 represented by the
represented by the DiffSSBasedOptimizableFunction returned by
getFunction(DataSet[], double[][]). As proposal distribution, a Gaussian distribution with given sampling
variance is used for each parameter.
Specifically, a new set of parameters 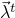 is drawn from a proposal distribution
is drawn from a proposal distribution 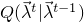 ,
where
,
where
![\[ Q(\vec{\lambda}^{t}|\vec{\lambda}^{t-1}) = \prod_{i} \mathcal{N}(\lambda_i^{t}|\lambda_i^{t},\sigma_i^2)\]](images/SamplingScoreBasedClassifier_LaTeXilb5_1.png)
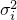 is the sampling variance for parameter
is the sampling variance for parameter 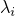 . The sampling variances are adapted to the
size of the event space of each parameter based on a class-dependent variance provided to the constructor. This adaption depends on the correct
implementation of
. The sampling variances are adapted to the
size of the event space of each parameter based on a class-dependent variance provided to the constructor. This adaption depends on the correct
implementation of DifferentiableStatisticalModel.getSizeOfEventSpaceForRandomVariablesOfParameter(int). Let 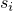 be the size of the event space
of the random variable of parameter
be the size of the event space
of the random variable of parameter 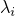 , and let
, and let 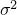 be the class-dependent variance for the
be the class-dependent variance for the SamplingDifferentiableStatisticalModel
that 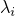 is a parameter of. Then
is a parameter of. Then 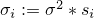 . If
. If 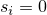 then
then 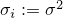 .
After a new set of parameters
.
After a new set of parameters 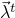 has been drawn, the sampling process decides if this new set of parameters is accepted
according to the distribution
has been drawn, the sampling process decides if this new set of parameters is accepted
according to the distribution 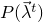 that we want to sample from.
Specifically, the parameters are accepted, iff.
that we want to sample from.
Specifically, the parameters are accepted, iff.
![\[ \alpha < \frac{ P(\vec{\lambda}^{t})Q(\vec{\lambda}^{t-1} | \vec{\lambda}^{t}) }{P(\vec{\lambda}^{t-1}) Q(\vec{\lambda}^{t} | \vec{\lambda}^{t-1})},\]](images/SamplingScoreBasedClassifier_LaTeXilb6_1.png)
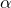 is drawn from a uniform distribution in
is drawn from a uniform distribution in ![$[0,1]$](images/SamplingScoreBasedClassifier_LaTeXil32_1.png) , i.e.
, i.e. Random.nextDouble().
Otherwise, the parameters are rejected and 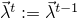 .
Since the Gaussian distribution is symmetric around its mean,
.
Since the Gaussian distribution is symmetric around its mean, 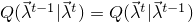 , both terms
cancel, and the acceptance probability depends only on the current and previous values of
, both terms
cancel, and the acceptance probability depends only on the current and previous values of 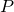 .
All sampled parameters are stored to separate temporary files for each concurrent sampling run by an internal
.
All sampled parameters are stored to separate temporary files for each concurrent sampling run by an internal SamplingComponent. The contents of these files
are stored together with the remaining representation of the SamplingScoreBasedClassifier, if AbstractClassifier.toXML() is called, and, hence,
can be stored to a monolithic file containing all information for, e.g., later classification procedures.
For determining the length of the burn-in phase and, as a consequence, the beginning of the stationary phase, a BurnInTest can be provided to the constructor of the classifier.
| Nested Class Summary | |
|---|---|
protected class |
SamplingScoreBasedClassifier.DiffSMSamplingComponent
The SamplingComponent that handles storing and loading sampled parameters values
to and from files. |
static class |
SamplingScoreBasedClassifier.SamplingScheme
Sampling scheme for sampling the parameters of the scoring functions. |
| Nested classes/interfaces inherited from class de.jstacs.classifiers.AbstractScoreBasedClassifier |
|---|
AbstractScoreBasedClassifier.DoubleTableResult |
| Field Summary | |
|---|---|
protected BurnInTest |
burnInTest
The BurnInTest, may be null for no test |
protected double[] |
currentParameters
the currently accepted parameters |
protected double |
currentScore
The score achieved using currentParameters |
protected double[] |
initParameters
The initial parameters if set by setInitParameters(double[]), null otherwise |
protected double[][] |
lastParameters
The last accepted parameters for all samplings, backup for iterative sampling when checking for BurnInTest |
protected double[] |
lastScore
The scores yielded for the parameters in lastParameters |
protected SamplingScoreBasedClassifierParameterSet |
params
Parameters |
protected double[] |
previousParameters
The previously accepted parameters, backup for rollbacks |
protected SamplingDifferentiableStatisticalModel[] |
scoringFunctions
SamplingDifferentiableStatisticalModels |
| Constructor Summary | |
|---|---|
protected |
SamplingScoreBasedClassifier(SamplingScoreBasedClassifierParameterSet params,
BurnInTest burnInTest,
double[] classVariances,
SamplingDifferentiableStatisticalModel... scoringFunctions)
Creates a new SamplingScoreBasedClassifier using the parameters in params,
a specified BurnInTest (or null for no burn-in test), a set of sampling variances,
which may be different for each of the classes (in analogy to equivalent sample size for the Dirichlet distribution),
and set set of SamplingDifferentiableStatisticalModels for each of the classes. |
|
SamplingScoreBasedClassifier(StringBuffer xml)
This is the constructor for Storable. |
| Method Summary | |
|---|---|
protected double |
doOneSamplingStep(DiffSSBasedOptimizableFunction function,
SamplingScoreBasedClassifier.SamplingScheme scheme,
double previousValue)
Performs one sampling step, i.e., one sampling of all parameter values. |
void |
doSingleSampling(DataSet[] s,
double[][] weights,
int numSteps,
String outfilePrefix)
Does a single sampling run for a predefined number of steps. |
protected void |
extractFurtherClassifierInfosFromXML(StringBuffer xml)
Extracts further information of a classifier from an XML representation. |
protected double[] |
getBestParameters()
Returns the sampled parameter values with the maximum value of the objective function |
CategoricalResult[] |
getClassifierAnnotation()
Returns an array of Results of dimension
AbstractClassifier.getNumberOfClasses() that contains information about the
classifier and for each class.
res[0] = new CategoricalResult( "classifier", "the kind of classifier", getInstanceName() ); |
boolean |
getDeleteOnExit()
Returns true if the temporary parameter files shall
be deleted on exit of the program. |
protected abstract DiffSSBasedOptimizableFunction |
getFunction(DataSet[] data,
double[][] weights)
Returns the function that should be sampled from. |
protected StringBuffer |
getFurtherClassifierInfos()
This method returns further information of a classifier as a StringBuffer. |
String |
getInstanceName()
Returns a short description of the classifier. |
protected double[] |
getMeanParameters(boolean testBurnIn,
int minBurnInSteps)
Returns the mean parameters over all samplings of all stationary phases. |
NumericalResultSet |
getNumericalCharacteristics()
Returns the subset of numerical values that are also returned by AbstractClassifier.getCharacteristics(). |
protected SamplingScoreBasedClassifier.DiffSMSamplingComponent |
getSamplingComponent()
Returns a sampling component suited for this SamplingScoreBasedClassifier |
protected double |
getScore(Sequence seq,
int cls,
boolean check)
This method returns the score for a given Sequence and a given
class. |
double[] |
getScores(DataSet s)
This method returns the scores of the classifier for any Sequence
in the DataSet. |
File |
getTempDir()
Returns the directory for parameter files set in this SamplingScoreBasedClassifier. |
protected void |
init(int starts,
boolean adaptVariance,
String outfilePrefix)
Initializes all internal fields and initializes the scoringFunctionss randomly |
boolean |
isInitialized()
This method gives information about the state of the classifier. |
void |
joinAndSetParameterFiles(boolean add,
File... files)
Combines parameter files such that they are accepted as parameter files of this SamplingScoreBasedClassifier |
protected double |
modifyFunctionValue(double value)
Allows for a modification of the value returned by the function obtained by getFunction(DataSet[], double[][]). |
protected void |
precomputeBurnInLength(SamplingScoreBasedClassifier.DiffSMSamplingComponent sfsc)
Precomputes the length of the burn-in phase, e.g. |
protected void |
sample(SamplingScoreBasedClassifier.DiffSMSamplingComponent sfsc,
DiffSSBasedOptimizableFunction function)
Samples as many steps as needed to get into the stationary phase according to burnInTest and then samples the number of
stationary steps as set in params. |
protected double |
sampleNSteps(DiffSSBasedOptimizableFunction function,
SamplingScoreBasedClassifier.DiffSMSamplingComponent component,
BurnInTest test,
int numSteps,
SamplingScoreBasedClassifier.SamplingScheme scheme)
Samples a predefined number of steps appended to the current sampling |
void |
setDeleteOnExit(boolean deleteOnExit)
If set to true (which is the default), the temporary files for storing sampled parameter
values are deleted on exit of the program. |
void |
setInitParameters(double[] parameters)
Sets the initial parameters of the sampling to parameters. |
void |
setTempDir(File tempDir)
Sets the directory for parameter files set in this SamplingScoreBasedClassifier. |
void |
train(DataSet[] s,
double[][] weights)
This method trains a classifier over an array of weighted DataSet
s. |
| Methods inherited from class de.jstacs.classifiers.AbstractScoreBasedClassifier |
|---|
check, check, classify, classify, clone, createDefaultClassWeights, getClassWeight, getClassWeights, getMultiClassScores, getNumberOfClasses, getPValue, getPValue, getResults, getScore, setClassWeights, setClassWeights, setThresholdClassWeights |
| Methods inherited from class de.jstacs.classifiers.AbstractClassifier |
|---|
classify, evaluate, evaluate, getAlphabetContainer, getCharacteristics, getLength, getXMLTag, toXML, train |
| Methods inherited from class java.lang.Object |
|---|
equals, finalize, getClass, hashCode, notify, notifyAll, toString, wait, wait, wait |
| Field Detail |
|---|
protected SamplingScoreBasedClassifierParameterSet params
protected SamplingDifferentiableStatisticalModel[] scoringFunctions
SamplingDifferentiableStatisticalModels
protected double[] currentParameters
protected double[] initParameters
setInitParameters(double[]), null otherwise
protected double currentScore
currentParameters
protected double[] previousParameters
protected double[][] lastParameters
BurnInTest
protected double[] lastScore
lastParameters
protected BurnInTest burnInTest
BurnInTest, may be null for no test
| Constructor Detail |
|---|
public SamplingScoreBasedClassifier(StringBuffer xml)
throws NonParsableException
Storable.
xml - the xml representation
NonParsableException - if the representation could not be parsed.
protected SamplingScoreBasedClassifier(SamplingScoreBasedClassifierParameterSet params,
BurnInTest burnInTest,
double[] classVariances,
SamplingDifferentiableStatisticalModel... scoringFunctions)
throws CloneNotSupportedException
SamplingScoreBasedClassifier using the parameters in params,
a specified BurnInTest (or null for no burn-in test), a set of sampling variances,
which may be different for each of the classes (in analogy to equivalent sample size for the Dirichlet distribution),
and set set of SamplingDifferentiableStatisticalModels for each of the classes.
params - the external parameters of this classifierburnInTest - the burn-in test (or null for no burn-in test)classVariances - the variances used for sampling for the parameters of each classscoringFunctions - the scoring functions for each of the classes
CloneNotSupportedException - if the scoring functions or the burn-in test could not be clonedVarianceRatioBurnInTest| Method Detail |
|---|
protected StringBuffer getFurtherClassifierInfos()
AbstractClassifierStringBuffer. This method is used by the method AbstractClassifier.toXML()
and should not be made public.
getFurtherClassifierInfos in class AbstractScoreBasedClassifierStringBufferAbstractClassifier.toXML()
protected void extractFurtherClassifierInfosFromXML(StringBuffer xml)
throws NonParsableException
AbstractClassifierAbstractClassifier.fromXML(StringBuffer) and
should not be made public.
extractFurtherClassifierInfosFromXML in class AbstractScoreBasedClassifierxml - the XML representation as StringBuffer
NonParsableException - if the information could not be parsed out of the XML
representation (the StringBuffer could not be parsed)AbstractClassifier.fromXML(StringBuffer)public CategoricalResult[] getClassifierAnnotation()
AbstractClassifierResults of dimension
AbstractClassifier.getNumberOfClasses() that contains information about the
classifier and for each class.
res[0] = new CategoricalResult( "classifier", "the kind of classifier", getInstanceName() );
res[1] = new CategoricalResult( "class info 0", "some information about the class", "info0" );
res[2] = new CategoricalResult( "class info 1", "some information about the class", "info1" );
...
getClassifierAnnotation in class AbstractClassifierResults that contains information about the
classifier
public NumericalResultSet getNumericalCharacteristics()
throws Exception
AbstractClassifierAbstractClassifier.getCharacteristics().
getNumericalCharacteristics in class AbstractClassifierException - if some of the characteristics could not be definedpublic String getInstanceName()
AbstractClassifier
getInstanceName in class AbstractClassifier
protected abstract DiffSSBasedOptimizableFunction getFunction(DataSet[] data,
double[][] weights)
throws Exception
data - the samplesweights - the weights of the sequences of the samples
Exception - if the function could not be createdprotected double modifyFunctionValue(double value)
getFunction(DataSet[], double[][]).
This is for instance necessary in case of LogGenDisMixFunction to
obtain a proper posterior or supervised posterior.
value - the original value
protected SamplingScoreBasedClassifier.DiffSMSamplingComponent getSamplingComponent()
SamplingScoreBasedClassifier
public File getTempDir()
SamplingScoreBasedClassifier.
If this value is null, the default directory of the executing OS is used for the parameter
files.
public void setTempDir(File tempDir)
SamplingScoreBasedClassifier.
If tempDir is null, the default directory of the executing OS is used for the parameter
files. If this value is reset after training, all sampled parameters will be lost.
The value set by this method is not stored in the XML-representation.
tempDir - the temp directorypublic boolean getDeleteOnExit()
true if the temporary parameter files shall
be deleted on exit of the program.
public void setDeleteOnExit(boolean deleteOnExit)
throws Exception
true (which is the default), the temporary files for storing sampled parameter
values are deleted on exit of the program. If this value is set to true it cannot be
reset to false, again, after sampling started due to the restrictions of File.deleteOnExit().
If you want to retain those
parameters, nonetheless, you can call AbstractClassifier.toXML()
and save this StringBuffer, which also contains the sampled
parameter values, somewhere.
The value set by this method is not stored in the XML-representation.
deleteOnExit - if temp files shall be deleted on exit
Exception - if set to false after sampling started
protected void init(int starts,
boolean adaptVariance,
String outfilePrefix)
throws Exception
scoringFunctionss randomly
starts - number of startsadaptVariance - if true, variance is adapted to size of event spaceoutfilePrefix - the prefix of the outfiles
Exception - if the scoring functions could not be initialized
protected double sampleNSteps(DiffSSBasedOptimizableFunction function,
SamplingScoreBasedClassifier.DiffSMSamplingComponent component,
BurnInTest test,
int numSteps,
SamplingScoreBasedClassifier.SamplingScheme scheme)
throws Exception
function - the objective functioncomponent - the sampling component with selected samplingtest - the burn-in testnumSteps - the number of stepsscheme - the SamplingScoreBasedClassifier.SamplingScheme
Exception - if either the function could not be evaluated on the current parameters or the
sampled parameters could not be stored
protected void sample(SamplingScoreBasedClassifier.DiffSMSamplingComponent sfsc,
DiffSSBasedOptimizableFunction function)
throws Exception
burnInTest and then samples the number of
stationary steps as set in params.
sfsc - the current sampling componentfunction - the objective function
Exception - if the sampling could not be extended, e.g. due to evaluation errors
protected double doOneSamplingStep(DiffSSBasedOptimizableFunction function,
SamplingScoreBasedClassifier.SamplingScheme scheme,
double previousValue)
throws Exception
function - the objective functionscheme - the SamplingScoreBasedClassifier.SamplingSchemepreviousValue - the value of the last sampling or minus infinity
for the first sampling run
Double.NaN if none
of the sampled parameters where accepted
Exception - if the function could not be evaluated or an unknown SamplingScoreBasedClassifier.SamplingScheme was provided
protected double getScore(Sequence seq,
int cls,
boolean check)
throws IllegalArgumentException,
NotTrainedException,
Exception
AbstractScoreBasedClassifierSequence and a given
class.
getScore in class AbstractScoreBasedClassifierseq - the Sequencecls - the index of the classcheck - the switch to decide whether to check
AlphabetContainer and the length of the
Sequence or not
Sequence and a given class
IllegalArgumentException - if something is wrong with the Sequence
seq
NotTrainedException - if the classifier is not trained
Exception - if something went wrong
public double[] getScores(DataSet s)
throws Exception
AbstractScoreBasedClassifierSequence
in the DataSet. The scores are stored in the array according to
the index of the Sequence in the DataSet.
getScores in class AbstractScoreBasedClassifiers - the DataSet
Exception - if something went wrongpublic void setInitParameters(double[] parameters)
parameters.
parameters - the initial parameterspublic boolean isInitialized()
AbstractClassifier
isInitialized in class AbstractClassifiertrue if the classifier is initialized and therefore able
to classify sequences, otherwise false
public void doSingleSampling(DataSet[] s,
double[][] weights,
int numSteps,
String outfilePrefix)
throws Exception
s - the dataweights - the weights for the datanumSteps - the number of sampling stepsoutfilePrefix - the prefix of the outfile where the parameter values
are stored
Exception - if the scoring functions could not be initialized or the sampling could not be extended, e.g. due to evaluation errors
public void train(DataSet[] s,
double[][] weights)
throws Exception
AbstractClassifierDataSet
s. That is why the following has to be fulfilled:
s.length == weights.length
weights[i] == null || s[i].getNumberOfElements() == weights[i].length.
AbstractClassifier.train(DataSet...).
DataSets are defined over the
underlying alphabet and length.
train in class AbstractClassifiers - an array of DataSetsweights - the weights for the DataSets
Exception - if the weights are incorrect or the training did not succeedAbstractClassifier.train(DataSet...)
protected void precomputeBurnInLength(SamplingScoreBasedClassifier.DiffSMSamplingComponent sfsc)
throws Exception
sfsc - the current sampling component
Exception - if the parameters values could not be parsed
protected double[] getBestParameters()
throws Exception
Exception - if the parameters values could not be parsed
protected double[] getMeanParameters(boolean testBurnIn,
int minBurnInSteps)
throws Exception
testBurnIn - true if the length of the burn-in phase shall be computedminBurnInSteps - minimum number of steps considered as burn-in
Exception - if the parameters values could not be parsed
public void joinAndSetParameterFiles(boolean add,
File... files)
throws Exception
SamplingScoreBasedClassifier
add - if true, parameter files are appended to the current ones, i.e., the number
of samplings is augmented by these filesfiles - the parameter files
- Throws:
Exception - if the parameter files could not be joined
|
||||||||||
| PREV CLASS NEXT CLASS | FRAMES NO FRAMES | |||||||||
| SUMMARY: NESTED | FIELD | CONSTR | METHOD | DETAIL: FIELD | CONSTR | METHOD | |||||||||