 de.jstacs.classifiers.AbstractClassifier
de.jstacs.classifiers.AbstractClassifier
 de.jstacs.classifiers.AbstractScoreBasedClassifier
de.jstacs.classifiers.AbstractScoreBasedClassifier
|
||||||||||
| PREV CLASS NEXT CLASS | FRAMES NO FRAMES | |||||||||
| SUMMARY: NESTED | FIELD | CONSTR | METHOD | DETAIL: FIELD | CONSTR | METHOD | |||||||||
java.lang.Objectde.jstacs.classifiers.AbstractClassifier
de.jstacs.classifiers.AbstractScoreBasedClassifier
public abstract class AbstractScoreBasedClassifier
This class is the main class for all score based classifiers. Score based classifiers enable you to compute many different measures easily. For instance one can use the package "ROCR" in R to compute or plot many of them.
| Nested Class Summary | |
|---|---|
static class |
AbstractScoreBasedClassifier.DoubleTableResult
This class is for Results given as a table of double
s. |
| Constructor Summary | |
|---|---|
AbstractScoreBasedClassifier(AlphabetContainer abc,
int classes)
The constructor for a homogeneous classifier. |
|
AbstractScoreBasedClassifier(AlphabetContainer abc,
int classes,
double classWeight)
The constructor for a homogeneous classifier. |
|
AbstractScoreBasedClassifier(AlphabetContainer abc,
int length,
int classes)
The constructor for an inhomogeneous classifier. |
|
AbstractScoreBasedClassifier(AlphabetContainer abc,
int length,
int classes,
double classWeight)
The constructor for an inhomogeneous classifier. |
|
AbstractScoreBasedClassifier(StringBuffer xml)
The standard constructor for the interface Storable. |
|
| Method Summary | |
|---|---|
protected void |
check(DataSet s)
This method checks if the given DataSet can be used. |
protected void |
check(Sequence seq)
This method checks if the given Sequence can be used. |
byte |
classify(Sequence seq)
This method classifies a sequence and returns the index i of
the class to which the sequence is assigned with
0 < i < getNumberOfClasses(). |
protected byte |
classify(Sequence seq,
boolean check)
This method classifies a Sequence. |
AbstractScoreBasedClassifier |
clone()
|
protected void |
createDefaultClassWeights(int classes,
double val)
This method creates new class weights. |
protected void |
extractFurtherClassifierInfosFromXML(StringBuffer xml)
Extracts further information of a classifier from an XML representation. |
protected double |
getClassWeight(int index)
Returns the class weight for the class with a given index. |
double[] |
getClassWeights()
Returns the specific class weights of a AbstractScoreBasedClassifier. |
protected StringBuffer |
getFurtherClassifierInfos()
This method returns further information of a classifier as a StringBuffer. |
protected double[][][] |
getMultiClassScores(DataSet[] s)
This method returns a multidimensional array with class specific scores. |
int |
getNumberOfClasses()
Returns the number of classes that can be distinguished. |
double[] |
getPValue(DataSet candidates,
DataSet bg)
Returns the p-values for all Sequences in the DataSet
candidates with respect to a given background DataSet
. |
double |
getPValue(Sequence candidate,
DataSet bg)
Returns the p-value for a Sequence candidate with
respect to a given background DataSet. |
protected boolean |
getResults(LinkedList list,
DataSet[] s,
double[][] weights,
AbstractPerformanceMeasureParameterSet<? extends PerformanceMeasure> params,
boolean exceptionIfNotComputeable)
This method computes the results for any evaluation of the classifier. |
double |
getScore(Sequence seq,
int i)
This method returns the score for a given Sequence and a given
class. |
protected abstract double |
getScore(Sequence seq,
int i,
boolean check)
This method returns the score for a given Sequence and a given
class. |
double[] |
getScores(DataSet s)
This method returns the scores of the classifier for any Sequence
in the DataSet. |
void |
setClassWeights(boolean add,
double... weights)
Sets new class weights. |
protected void |
setClassWeights(boolean add,
double[] weights,
int start)
Sets new class weights. |
void |
setThresholdClassWeights(boolean add,
double t)
Sets a new threshold for 2-class-classifiers. Only available if this AbstractScoreBasedClassifier distinguishes
between 2 classes 0 and 1. |
| Methods inherited from class de.jstacs.classifiers.AbstractClassifier |
|---|
classify, evaluate, evaluate, getAlphabetContainer, getCharacteristics, getClassifierAnnotation, getInstanceName, getLength, getNumericalCharacteristics, getXMLTag, isInitialized, toXML, train, train |
| Methods inherited from class java.lang.Object |
|---|
equals, finalize, getClass, hashCode, notify, notifyAll, toString, wait, wait, wait |
| Constructor Detail |
|---|
public AbstractScoreBasedClassifier(AlphabetContainer abc,
int classes)
0.
abc - the alphabets that are usedclasses - the number of different classesAbstractScoreBasedClassifier(AlphabetContainer,
int, int, double)
public AbstractScoreBasedClassifier(AlphabetContainer abc,
int classes,
double classWeight)
classWeight.
abc - the alphabets that are usedclasses - the number of different classesclassWeight - the value of all class weightsAbstractScoreBasedClassifier(AlphabetContainer,
int, int, double)
public AbstractScoreBasedClassifier(AlphabetContainer abc,
int length,
int classes)
0.
abc - the alphabets that are usedlength - the length of the sequences that can be classifiedclasses - the number of different classesAbstractScoreBasedClassifier(AlphabetContainer,
int, int, double)
public AbstractScoreBasedClassifier(AlphabetContainer abc,
int length,
int classes,
double classWeight)
throws IllegalArgumentException
classWeight.
abc - the alphabets that are usedlength - the length of the sequences that can be classifiedclasses - the number of different classesclassWeight - the value of all class weights
IllegalArgumentException - if the length and the possible length of the
AlphabetContainer does not match or the number of
classes is less than 2AbstractClassifier.AbstractClassifier(AlphabetContainer, int)
public AbstractScoreBasedClassifier(StringBuffer xml)
throws NonParsableException
Storable.
Creates a new AbstractScoreBasedClassifier out of its XML
representation.
xml - the XML representation as StringBuffer
NonParsableException - if the AbstractScoreBasedClassifier could not be
reconstructed out of the XML representation (the
StringBuffer could not be parsed)Storable,
AbstractClassifier.AbstractClassifier(StringBuffer)| Method Detail |
|---|
public AbstractScoreBasedClassifier clone()
throws CloneNotSupportedException
clone in class AbstractClassifierCloneNotSupportedException
public byte classify(Sequence seq)
throws Exception
AbstractClassifieri of
the class to which the sequence is assigned with
0 < i < getNumberOfClasses().
classify in class AbstractClassifierseq - the sequence to be classified
Exception - if the classifier is not trained or something is wrong with
the sequence
protected double[][][] getMultiClassScores(DataSet[] s)
throws Exception
AbstractClassifierresult[d][n][c] returns the score of class c for sequence n of the data set s[d]. The class with the maximum score
for any sequence is the predicted class of the sequence.
getMultiClassScores in class AbstractClassifiers - the data sets
Exception - if the scores can not be computedAbstractClassifier.getResults(LinkedList, DataSet[], double[][], AbstractPerformanceMeasureParameterSet, boolean)
protected boolean getResults(LinkedList list,
DataSet[] s,
double[][] weights,
AbstractPerformanceMeasureParameterSet<? extends PerformanceMeasure> params,
boolean exceptionIfNotComputeable)
throws Exception
AbstractClassifier
getResults in class AbstractClassifierlist - a list adding the resultss - the array of DataSetsweights - the weights of the sequences for each data setparams - the current parametersexceptionIfNotComputeable - indicates the method throws an Exception if a measure
could not be computed
Exception - if something went wrongAbstractClassifier.evaluate(AbstractPerformanceMeasureParameterSet, boolean, DataSet[], double[][]),
NumericalResult,
Resultpublic double[] getClassWeights()
AbstractScoreBasedClassifier.
public int getNumberOfClasses()
AbstractClassifier
getNumberOfClasses in class AbstractClassifier
public double getScore(Sequence seq,
int i)
throws Exception
Sequence and a given
class.
seq - the given Sequencei - the index of the class
Sequence and a given class
Exception - if something went wronggetScore(Sequence, int, boolean)
public final void setClassWeights(boolean add,
double... weights)
throws ClassDimensionException
i given class 0 to
class n are computed to classify this item into a class. The
class weights are added to each of these logarithmic probabilities. As
higher (relational) the class weight of class j, as more
probable it becomes, that any item is classified into this class. 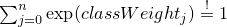 ,
the class weights may be interpreted as logarithmic class-a-priori-probabilities.
,
the class weights may be interpreted as logarithmic class-a-priori-probabilities.
add - indicates if the class weights are added to the current class
weightsweights - the array of weights, for each class the weight that is added
in a classification
ClassDimensionException - if something is wrong with the number of classes
protected final void setClassWeights(boolean add,
double[] weights,
int start)
add - indicates if the class weights are added to the current class
weightsweights - an array of weights that might have more entries than the classifier has classesstart - the start indexsetClassWeights(boolean, double...)
public final void setThresholdClassWeights(boolean add,
double t)
throws OperationNotSupportedException
AbstractScoreBasedClassifier distinguishes
between 2 classes 0 and 1. In this case, t will be interpreted as
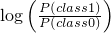 . A large
. A large
t (greater than 0) makes the classifier to decide more often for
class 1. A small t (smaller than 0) makes the classifier to
decide more often for class 0.
add - indicates if the class weights are added to the current class
weightst - the new threshold
OperationNotSupportedException - if the classifier is no 2-class-classifierprotected StringBuffer getFurtherClassifierInfos()
AbstractClassifierStringBuffer. This method is used by the method AbstractClassifier.toXML()
and should not be made public.
getFurtherClassifierInfos in class AbstractClassifierStringBufferAbstractClassifier.toXML()
protected void check(DataSet s)
throws NotTrainedException,
IllegalArgumentException
DataSet can be used.
s - the DataSet to be checked
NotTrainedException - if the classifier is not trained
IllegalArgumentException - if something is wrong with the DataSet sAlphabetContainer.checkConsistency(AlphabetContainer)
protected void check(Sequence seq)
throws NotTrainedException,
IllegalArgumentException
Sequence can be used.
seq - the Sequence to be checked
NotTrainedException - if the classifier is not trained
IllegalArgumentException - if something is wrong with the Sequence
seq
protected byte classify(Sequence seq,
boolean check)
throws Exception
Sequence. It enables you to check the
constraints (alphabets, length,
AbstractClassifier.isInitialized() ).
seq - the Sequencecheck - indicates if the constraints will be checked
Sequence is assigned to
Exception - if something went wrongcheck(Sequence)
protected void createDefaultClassWeights(int classes,
double val)
throws IllegalArgumentException
val. So the class weights do not have any influence on
the classification.
classes - the number of different classesval - the value that is used for all classes
IllegalArgumentException - if the number of classes is below 2
protected void extractFurtherClassifierInfosFromXML(StringBuffer xml)
throws NonParsableException
AbstractClassifierAbstractClassifier.fromXML(StringBuffer) and
should not be made public.
extractFurtherClassifierInfosFromXML in class AbstractClassifierxml - the XML representation as StringBuffer
NonParsableException - if the information could not be parsed out of the XML
representation (the StringBuffer could not be parsed)AbstractClassifier.fromXML(StringBuffer)protected double getClassWeight(int index)
index.
index - the given index of the class
protected abstract double getScore(Sequence seq,
int i,
boolean check)
throws IllegalArgumentException,
NotTrainedException,
Exception
Sequence and a given
class.
seq - the Sequencei - the index of the classcheck - the switch to decide whether to check
AlphabetContainer and the length of the
Sequence or not
Sequence and a given class
NotTrainedException - if the classifier is not trained
IllegalArgumentException - if something is wrong with the Sequence
seq
Exception - if something went wrong
public double[] getScores(DataSet s)
throws Exception
Sequence
in the DataSet. The scores are stored in the array according to
the index of the Sequence in the DataSet.
s - the DataSet
Exception - if something went wrong
public double getPValue(Sequence candidate,
DataSet bg)
throws Exception
Sequence candidate with
respect to a given background DataSet.
Sequences that have a
score that is at least as high as the score for the
candidate.
for-loop. In
such cases one should use the method that works on two DataSets.
candidate - the candidate Sequencebg - the background DataSet
Sequence candidate
Exception - if something went wronggetPValue(DataSet, DataSet),
PValueComputation.getPValue(double[], double)
public double[] getPValue(DataSet candidates,
DataSet bg)
throws Exception
Sequences in the DataSet
candidates with respect to a given background DataSet
.
Sequences that have a
score that is at least as high as the score for a Sequence in
candidates.
candidates - the DataSet with candidate sequencesbg - the background data set
candidates
Exception - if something went wronggetPValue(Sequence, DataSet),
PValueComputation.getPValue(double[], double)
|
||||||||||
| PREV CLASS NEXT CLASS | FRAMES NO FRAMES | |||||||||
| SUMMARY: NESTED | FIELD | CONSTR | METHOD | DETAIL: FIELD | CONSTR | METHOD | |||||||||