de.jstacs.sequenceScores.statisticalModels.trainable.hmm.states.emissions
Interface DifferentiableEmission
- All Superinterfaces:
- Cloneable, Emission, Storable
- All Known Implementing Classes:
- AbstractConditionalDiscreteEmission, DiscreteEmission, GaussianEmission, PhyloDiscreteEmission, PluginGaussianEmission, ReferenceSequenceDiscreteEmission, SilentEmission, UniformEmission
public interface DifferentiableEmission
- extends Emission
This interface declares all methods needed in an emission during a numerical optimization of HMM.
- Author:
- Jens Keilwagen
|
Method Summary |
void |
addGradientOfLogPriorTerm(double[] grad,
int offset)
This method computes the gradient of Emission.getLogPriorTerm() for each
parameter of this model. |
void |
fillCurrentParameter(double[] params)
Fills the current parameters in the global code>params array using the internal offset. |
void |
fillSamplingGroups(int parameterOffset,
LinkedList<int[]> list)
Adds the groups of indexes of those parameters of this emission that should be sampled
together in one step of a grouped sampling procedure, each as an int[], into list. |
double |
getLogProbAndPartialDerivationFor(boolean forward,
int startPos,
int endPos,
IntList indices,
DoubleList partDer,
Sequence seq)
Returns the logarithmic score for a Sequence beginning at
position start in the Sequence and fills lists with
the indices and the partial derivations. |
int |
getNumberOfParameters()
Returns the number of parameters of this emission. |
int |
getSizeOfEventSpace()
Returns the size of the event space, i.e., the number of possible outcomes,
for the random variables of this emission |
void |
setParameter(double[] params,
int offset)
This method sets the internal parameters using the given global parameter array, the global offset of the HMM and the internal offset. |
int |
setParameterOffset(int offset)
This method sets the internal parameter offset and returns the new parameter offset for further use. |
| Methods inherited from interface de.jstacs.sequenceScores.statisticalModels.trainable.hmm.states.emissions.Emission |
addToStatistic, estimateFromStatistic, getAlphabetContainer, getLogPriorTerm, getLogProbFor, getNodeLabel, getNodeShape, initializeFunctionRandomly, joinStatistics, resetStatistic, setParameters, toString |
fillCurrentParameter
void fillCurrentParameter(double[] params)
- Fills the current parameters in the global code>params array using the internal offset.
- Parameters:
params - the global parameter array of the HMM- See Also:
setParameterOffset(int)
setParameter
void setParameter(double[] params,
int offset)
- This method sets the internal parameters using the given global parameter array, the global offset of the HMM and the internal offset.
- Parameters:
params - the global parameter array of the classifieroffset - the offset of the HMM- See Also:
setParameterOffset(int)
setParameterOffset
int setParameterOffset(int offset)
- This method sets the internal parameter offset and returns the new parameter offset for further use.
- Parameters:
offset - the offset to be set
- Returns:
- the new parameter offset
getLogProbAndPartialDerivationFor
double getLogProbAndPartialDerivationFor(boolean forward,
int startPos,
int endPos,
IntList indices,
DoubleList partDer,
Sequence seq)
throws OperationNotSupportedException
- Returns the logarithmic score for a
Sequence beginning at
position start in the Sequence and fills lists with
the indices and the partial derivations.
- Parameters:
forward - a switch whether to use the forward or the reverse complementary strand of the sequenceseq - the SequencestartPos - the start position in the SequenceendPos - the end position in the Sequenceindices - an IntList of indices, after method invocation the
list should contain the indices i where
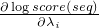 is not zero
is not zeropartDer - a DoubleList of partial derivations, after method
invocation the list should contain the corresponding
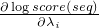 that are not zero
that are not zero
- Returns:
- the logarithmic score for the
Sequence
- Throws:
OperationNotSupportedException - if forward==false and the reverse complement of the sequence can not be computed
addGradientOfLogPriorTerm
void addGradientOfLogPriorTerm(double[] grad,
int offset)
- This method computes the gradient of
Emission.getLogPriorTerm() for each
parameter of this model. The results are added to the array
grad beginning at index (offset + internal offset).
- Parameters:
grad - the array of gradientsoffset - the start index of the HMM in the grad array, where the
partial derivations for the parameters of the HMM shall be
entered- See Also:
Emission.getLogPriorTerm(),
setParameterOffset(int)
fillSamplingGroups
void fillSamplingGroups(int parameterOffset,
LinkedList<int[]> list)
- Adds the groups of indexes of those parameters of this emission that should be sampled
together in one step of a grouped sampling procedure, each as an
int[], into list.
In most cases, one group should contain the parameters that are living on a common simplex.
The internal indexes of the parameters are incremeneted by an external parameterOffset
- Parameters:
parameterOffset - the external parameter offsetlist - the list of sampling groups
getNumberOfParameters
int getNumberOfParameters()
- Returns the number of parameters of this emission.
- Returns:
- the number of parameters
getSizeOfEventSpace
int getSizeOfEventSpace()
- Returns the size of the event space, i.e., the number of possible outcomes,
for the random variables of this emission
- Returns:
- the size of the event space
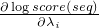 is not zero
is not zero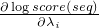 that are not zero
that are not zero