 de.jstacs.sequenceScores.statisticalModels.trainable.hmm.states.emissions.SilentEmission
de.jstacs.sequenceScores.statisticalModels.trainable.hmm.states.emissions.SilentEmission
|
||||||||||
| PREV CLASS NEXT CLASS | FRAMES NO FRAMES | |||||||||
| SUMMARY: NESTED | FIELD | CONSTR | METHOD | DETAIL: FIELD | CONSTR | METHOD | |||||||||
java.lang.Objectde.jstacs.sequenceScores.statisticalModels.trainable.hmm.states.emissions.SilentEmission
public final class SilentEmission
This class implements a silent emission which is used to create silent states.
| Constructor Summary | |
|---|---|
SilentEmission()
The main constructor. |
|
SilentEmission(StringBuffer xml)
The standard constructor for the interface Storable. |
|
| Method Summary | |
|---|---|
void |
acceptParameters()
This methods accepts the drawn parameters. |
void |
addGradientOfLogPriorTerm(double[] gradient,
int offset)
This method computes the gradient of Emission.getLogPriorTerm() for each
parameter of this model. |
void |
addToStatistic(boolean forward,
int startPos,
int endPos,
double weight,
Sequence seq)
This method adds the weight to the internal sufficient statistic. |
SilentEmission |
clone()
|
void |
drawParametersFromStatistic()
This method draws the parameters using a sufficient statistic representing a posteriori density. |
void |
estimateFromStatistic()
This method estimates the parameters from the internal sufficient statistic. |
void |
extendSampling(int sampling,
boolean append)
This method allows to extend a sampling. |
void |
fillCurrentParameter(double[] params)
Fills the current parameters in the global code>params array using the internal offset. |
void |
fillSamplingGroups(int parameterOffset,
LinkedList<int[]> list)
Adds the groups of indexes of those parameters of this emission that should be sampled together in one step of a grouped sampling procedure, each as an int[], into list. |
AlphabetContainer |
getAlphabetContainer()
This method returns the AlphabetContainer of this emission. |
double |
getLogGammaScoreFromStatistic()
This method calculates a score for the current statistics, which is independent from the current parameters In general the gamma-score is a product of gamma-functions parameterized with the current statistics |
double |
getLogPosteriorFromStatistic()
This method calculates the a-posteriori probability for the current statistics |
double |
getLogPriorTerm()
Returns a value that is proportional to the log of the prior. |
double |
getLogProbAndPartialDerivationFor(boolean forward,
int startPos,
int endPos,
IntList indices,
DoubleList partDer,
Sequence seq)
Returns the logarithmic score for a Sequence beginning at
position start in the Sequence and fills lists with
the indices and the partial derivations. |
double |
getLogProbFor(boolean forward,
int startPos,
int endPos,
Sequence seq)
This method computes the logarithm of the likelihood. |
String |
getNodeLabel(double weight,
String name,
NumberFormat nf)
Returns the graphviz label of the node containing this emission. |
String |
getNodeShape(boolean forward)
Returns the graphviz string for the shape of the node. |
int |
getNumberOfParameters()
Returns the number of parameters of this emission. |
int |
getSizeOfEventSpace()
Returns the size of the event space, i.e., the number of possible outcomes, for the random variables of this emission |
void |
initForSampling(int starts)
This method initializes the instance for the sampling. |
void |
initializeFunctionRandomly()
This method initializes the emission randomly. |
boolean |
isInSamplingMode()
This method returns true if the object is currently used in
a sampling, otherwise false. |
void |
joinStatistics(Emission... emissions)
This method joins the statistics of different instances and sets this joined statistic as statistic of each instance. |
boolean |
parseNextParameterSet()
This method allows the user to parse the next set of parameters (from a file). |
boolean |
parseParameterSet(int sampling,
int n)
This method allows the user to parse the set of parameters with index n of a certain sampling (from a file). |
void |
resetStatistic()
This method resets the internal sufficient statistic. |
void |
samplingStopped()
This method is the opposite of the method SamplingComponent.extendSampling(int, boolean). |
void |
setParameter(double[] params,
int offset)
This method sets the internal parameters using the given global parameter array, the global offset of the HMM and the internal offset. |
int |
setParameterOffset(int offset)
This method sets the internal parameter offset and returns the new parameter offset for further use. |
void |
setParameters(Emission t)
Set values of parameters of the instance to the value of the parameters of the given instance. |
String |
toString(NumberFormat nf)
This method returns a String representation of the instance. |
StringBuffer |
toXML()
This method returns an XML representation as StringBuffer of an
instance of the implementing class. |
| Methods inherited from class java.lang.Object |
|---|
equals, finalize, getClass, hashCode, notify, notifyAll, toString, wait, wait, wait |
| Constructor Detail |
|---|
public SilentEmission()
public SilentEmission(StringBuffer xml)
Storable.
Constructs a SilentEmission out of an XML representation.
xml - the XML representation as StringBuffer| Method Detail |
|---|
public SilentEmission clone()
throws CloneNotSupportedException
clone in class ObjectCloneNotSupportedExceptionpublic StringBuffer toXML()
StorableStringBuffer of an
instance of the implementing class.
toXML in interface Storable
public double getLogProbAndPartialDerivationFor(boolean forward,
int startPos,
int endPos,
IntList indices,
DoubleList partDer,
Sequence seq)
throws OperationNotSupportedException
DifferentiableEmissionSequence beginning at
position start in the Sequence and fills lists with
the indices and the partial derivations.
getLogProbAndPartialDerivationFor in interface DifferentiableEmissionforward - a switch whether to use the forward or the reverse complementary strand of the sequencestartPos - the start position in the SequenceendPos - the end position in the Sequenceindices - an IntList of indices, after method invocation the
list should contain the indices i where
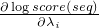 is not zero
is not zeropartDer - a DoubleList of partial derivations, after method
invocation the list should contain the corresponding
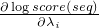 that are not zero
that are not zeroseq - the Sequence
Sequence
OperationNotSupportedException - if forward==false and the reverse complement of the sequence can not be computed
public double getLogProbFor(boolean forward,
int startPos,
int endPos,
Sequence seq)
throws OperationNotSupportedException
Emission
getLogProbFor in interface Emissionforward - whether to use the forward or the reverse strandstartPos - the start positionendPos - the end positionseq - the sequence
OperationNotSupportedException - if forward=false and the reverse complement of the sequence seq is not definedpublic void joinStatistics(Emission... emissions)
Emission
joinStatistics in interface Emissionemissions - the emissions to be joined
public void addToStatistic(boolean forward,
int startPos,
int endPos,
double weight,
Sequence seq)
throws OperationNotSupportedException
Emissionweight to the internal sufficient statistic.
addToStatistic in interface Emissionforward - whether to use the forward or the reverse strandstartPos - the start positionendPos - the end positionweight - the weight of the sequenceseq - the sequence
OperationNotSupportedException - if forward=false and the reverse complement of the sequence seq is not definedpublic void estimateFromStatistic()
Emission
estimateFromStatistic in interface Emissionpublic void resetStatistic()
Emission
resetStatistic in interface Emissionpublic AlphabetContainer getAlphabetContainer()
EmissionAlphabetContainer of this emission.
getAlphabetContainer in interface EmissionAlphabetContainer of this emission
public void addGradientOfLogPriorTerm(double[] gradient,
int offset)
DifferentiableEmissionEmission.getLogPriorTerm() for each
parameter of this model. The results are added to the array
grad beginning at index (offset + internal offset).
addGradientOfLogPriorTerm in interface DifferentiableEmissiongradient - the array of gradientsoffset - the start index of the HMM in the grad array, where the
partial derivations for the parameters of the HMM shall be
enteredEmission.getLogPriorTerm(),
DifferentiableEmission.setParameterOffset(int)public double getLogPriorTerm()
Emission
getLogPriorTerm in interface EmissionStatisticalModel.getLogPriorTerm()public void fillCurrentParameter(double[] params)
DifferentiableEmission
fillCurrentParameter in interface DifferentiableEmissionparams - the global parameter array of the HMMDifferentiableEmission.setParameterOffset(int)
public void setParameter(double[] params,
int offset)
DifferentiableEmission
setParameter in interface DifferentiableEmissionparams - the global parameter array of the classifieroffset - the offset of the HMMDifferentiableEmission.setParameterOffset(int)public int setParameterOffset(int offset)
DifferentiableEmission
setParameterOffset in interface DifferentiableEmissionoffset - the offset to be set
public void initializeFunctionRandomly()
Emission
initializeFunctionRandomly in interface Emissionpublic void drawParametersFromStatistic()
SamplingFromStatisticSamplingComponent.acceptParameters() so that they can later be parsed using the
methods of the interface.
SamplingComponent.initForSampling(int) should be
called.
drawParametersFromStatistic in interface SamplingFromStatisticSamplingComponent.initForSampling(int),
SamplingComponent.acceptParameters()
public void extendSampling(int sampling,
boolean append)
throws IOException
SamplingComponent
extendSampling in interface SamplingComponentsampling - the index of the samplingappend - whether to append the sampled parameters to an existing file
or to overwrite the file
IOException - if the file could not be handled correctly
public void initForSampling(int starts)
throws IOException
SamplingComponent
initForSampling in interface SamplingComponentstarts - the number of different sampling starts that will be done
IOException - if something went wrongFile.createTempFile(String, String, java.io.File )public boolean isInSamplingMode()
SamplingComponenttrue if the object is currently used in
a sampling, otherwise false.
isInSamplingMode in interface SamplingComponenttrue if the object is currently used in a sampling,
otherwise falsepublic boolean parseNextParameterSet()
SamplingComponent
parseNextParameterSet in interface SamplingComponenttrue if the parameters could be parsed, otherwise
falseSamplingComponent.parseParameterSet(int, int)
public boolean parseParameterSet(int sampling,
int n)
throws Exception
SamplingComponentn of a certain sampling (from a file). The
internal numbering should start with 0. The parameter set with index 0 is
the initial (random) parameter set. It is recommended that a series of
parameter sets is accessed by the following lines:
for( sampling = 0; sampling < numSampling; sampling++ )
{
while( b )
{
//do something
b = parseNextParameterSet();
}
parseParameterSet in interface SamplingComponentsampling - the index of the samplingn - the index of the parameter set
true if the parameter set could be parsed
Exception - if there is a problem with parsing the parametersSamplingComponent.parseNextParameterSet()
public void samplingStopped()
throws IOException
SamplingComponentSamplingComponent.extendSampling(int, boolean). It can be
used for closing any streams of writer, ...
samplingStopped in interface SamplingComponentIOException - if something went wrongSamplingComponent.extendSampling(int, boolean)
public void acceptParameters()
throws IOException
SamplingComponent
acceptParameters in interface SamplingComponentIOException - if the file could not be handled correctlypublic double getLogPosteriorFromStatistic()
SamplingFromStatistic
getLogPosteriorFromStatistic in interface SamplingFromStatisticpublic String toString(NumberFormat nf)
EmissionString representation of the instance.
toString in interface Emissionnf - the NumberFormat for the String representation of parameters or probabilities
String representation of the instancepublic double getLogGammaScoreFromStatistic()
SamplingEmission
getLogGammaScoreFromStatistic in interface SamplingEmissionpublic String getNodeShape(boolean forward)
Emission
getNodeShape in interface Emissionforward - if this emission is used on the forward strand
public String getNodeLabel(double weight,
String name,
NumberFormat nf)
Emission
getNodeLabel in interface Emissionweight - the weight of the node which is represented by
the color of the node, or -1 for no representation, i.e.,
white backgroundname - the name of the state using this emissionnf - the NumberFormat for formatting the textual representation of this emission
public void fillSamplingGroups(int parameterOffset,
LinkedList<int[]> list)
DifferentiableEmissionint[], into list.
In most cases, one group should contain the parameters that are living on a common simplex.
The internal indexes of the parameters are incremeneted by an external parameterOffset
fillSamplingGroups in interface DifferentiableEmissionparameterOffset - the external parameter offsetlist - the list of sampling groupspublic int getNumberOfParameters()
DifferentiableEmission
getNumberOfParameters in interface DifferentiableEmissionpublic int getSizeOfEventSpace()
DifferentiableEmission
getSizeOfEventSpace in interface DifferentiableEmission
public void setParameters(Emission t)
throws IllegalArgumentException
Emission
setParameters in interface Emissiont - the emission with the parameters to be set
IllegalArgumentException - if the assumption about the same class for given and current instance is wrong
|
||||||||||
| PREV CLASS NEXT CLASS | FRAMES NO FRAMES | |||||||||
| SUMMARY: NESTED | FIELD | CONSTR | METHOD | DETAIL: FIELD | CONSTR | METHOD | |||||||||