 de.jstacs.sequenceScores.statisticalModels.trainable.AbstractTrainableStatisticalModel
de.jstacs.sequenceScores.statisticalModels.trainable.AbstractTrainableStatisticalModel
 de.jstacs.sequenceScores.statisticalModels.trainable.mixture.AbstractMixtureTrainSM
de.jstacs.sequenceScores.statisticalModels.trainable.mixture.AbstractMixtureTrainSM
|
||||||||||
| PREV CLASS NEXT CLASS | FRAMES NO FRAMES | |||||||||
| SUMMARY: NESTED | FIELD | CONSTR | METHOD | DETAIL: FIELD | CONSTR | METHOD | |||||||||
java.lang.Objectde.jstacs.sequenceScores.statisticalModels.trainable.AbstractTrainableStatisticalModel
de.jstacs.sequenceScores.statisticalModels.trainable.mixture.AbstractMixtureTrainSM
public abstract class AbstractMixtureTrainSM
This is the abstract class for all kinds of mixture models. It enables the
user to train the parameters using AbstractMixtureTrainSM.Algorithm.EM or
AbstractMixtureTrainSM.Algorithm.GIBBS_SAMPLING. If this instance is trained using
AbstractMixtureTrainSM.Algorithm.GIBBS_SAMPLING the internal models that will be adjusted
have to implement SamplingComponent. If you use Gibbs sampling
temporary files will be created in the Java temp folder. These files will be
deleted if no reference to the current instance exists and the Garbage
Collector is called. Therefore it is recommended to call the Garbage
Collector explicitly at the end of any application.
The model stores a reference to the last data set used in train.
This enables the user to estimate the parameters iteratively beginning with
the current set of parameters. Therefore you can use the method
continueIterations(double[], double[][], int, int)
.
The method setOutputStream(OutputStream) enables
the user to get comments from the
train(DataSet, double[]) method or to repress
them.
The method getScoreForBestRun() enables the user
to optimize different instances of the same model (
clone()) using the EM-algorithm on different
CPUs, to compare the results and to select the best trained model. This might
be useful to get the results faster (measured in real time).
The reference to the internal data set is not stored if the model is stored
in a StringBuffer. So you can use these methods only after training
the parameters after (re)creating a model.
SamplingComponent,
System.gc()| Nested Class Summary | |
|---|---|
static class |
AbstractMixtureTrainSM.Algorithm
This enum defines the different types of algorithms that can
be used in an AbstractMixtureTrainSM. |
static class |
AbstractMixtureTrainSM.Parameterization
This enum defines the different types of parameterization
for a probability that can be used in an AbstractMixtureTrainSM. |
| Field Summary | |
|---|---|
protected AbstractMixtureTrainSM.Algorithm |
algorithm
The type of algorithm. |
protected boolean |
algorithmHasBeenRun
A switch which indicates that the algorithm for determining the parameters has been run. |
protected TrainableStatisticalModel[] |
alternativeModel
The alternative models for the EM. |
protected double |
best
This field contains the value of objective function of the best start of the training. |
protected BurnInTest |
burnInTest
The BurnInTest that is used to stop the sampling. |
protected double[] |
componentHyperParams
The hyperparameters for estimating the probabilities of the components. |
protected double[] |
compProb
This array is used while training to avoid creating many new objects. |
protected int[] |
counter
The current index of the parameter set while adjustment (optimization). |
protected int |
dimension
The number of dimensions. |
protected boolean |
estimateComponentProbs
The switch for estimating the component probabilities or not. |
protected File[] |
file
The file in which the component probabilities are stored. |
protected BufferedReader |
filereader
Reading component probabilities from a file. |
protected BufferedWriter |
filewriter
Saving component probabilities in a file. |
protected int |
initialIteration
The number of initial iterations. |
protected double[] |
logWeights
The log probabilities for each component. |
protected TrainableStatisticalModel[] |
model
The model for the sequences. |
protected boolean[] |
optimizeModel
A switch for each model whether to optimize/adjust or not. |
protected DataSet[] |
sample
The data set that was used in the last training. |
protected int |
samplingIndex
The current index of the sampling. |
protected double[][] |
seqWeights
The weights of the (sub-)sequence used to train the components (internal models). |
protected SafeOutputStream |
sostream
This is the stream for writing information while training. |
protected int |
starts
The number of starts. |
protected int |
stationaryIteration
The number of (stationary) iterations of the Gibbs Sampler. |
protected double[] |
weights
The probabilities for each component. |
| Fields inherited from class de.jstacs.sequenceScores.statisticalModels.trainable.AbstractTrainableStatisticalModel |
|---|
alphabets, length |
| Constructor Summary | |
|---|---|
protected |
AbstractMixtureTrainSM(int length,
TrainableStatisticalModel[] models,
boolean[] optimizeModel,
int dimension,
int starts,
boolean estimateComponentProbs,
double[] componentHyperParams,
double[] weights,
AbstractMixtureTrainSM.Algorithm algorithm,
double alpha,
TerminationCondition tc,
AbstractMixtureTrainSM.Parameterization parametrization,
int initialIteration,
int stationaryIteration,
BurnInTest burnInTest)
Creates a new AbstractMixtureTrainSM. |
protected |
AbstractMixtureTrainSM(StringBuffer xml)
The standard constructor for the interface Storable. |
| Method Summary | |
|---|---|
boolean |
algorithmHasBeenRun()
This method indicates whether the parameters of the model has been determined by the internal algorithm. |
protected void |
checkLength(int index,
int l)
This method checks if the length l of the model with index
index is capable for the current instance. |
protected void |
checkModelsForGibbsSampling()
This method can be used to check whether the necessary models have implemented the SamplingComponent. |
AbstractMixtureTrainSM |
clone()
Follows the conventions of Object's clone()-method. |
protected double |
continueIterations(double[] dataWeights,
double[][] seqweights)
This method will run the train algorithm for the current model on the internal data set. |
protected double |
continueIterations(double[] dataWeights,
double[][] seqweights,
int iterations,
int start)
This method will run the train algorithm for the current model on the internal sample. |
protected double[][] |
createSeqWeightsArray()
Creates an array that can be used for weighting sequences in the algorithm. |
protected double[][] |
doFirstIteration(DataSet data,
double[] dataWeights)
This method will do the first step in the train algorithm for the current model. |
protected double[][] |
doFirstIteration(DataSet data,
double[] dataWeights,
MultivariateRandomGenerator m,
MRGParams[] params)
This method will do the first step in the train algorithm for the current model. |
protected abstract double[][] |
doFirstIteration(double[] dataWeights,
MultivariateRandomGenerator m,
MRGParams[] params)
This method will do the first step in the train algorithm for the current model on the internal data set. |
static int |
draw(double[] w,
int start)
This method draws an index of an array corresponding to the probabilities encoded in the entries of the array. |
DataSet |
emitDataSet(int n,
int... lengths)
This method returns a DataSet object containing artificial
sequence(s). |
protected abstract Sequence[] |
emitDataSetUsingCurrentParameterSet(int n,
int... lengths)
The method returns an array of sequences using the current parameter set. |
protected void |
extendSampling(int sampling)
This method prepares the model to extend an existing sampling. |
protected void |
extractFurtherInformation(StringBuffer xml)
This method is used in the subclasses to extract further information from the XML representation and to set these as values of the instance. |
protected void |
finalize()
|
protected void |
fromXML(StringBuffer representation)
This method should only be used by the constructor that works on a StringBuffer. |
ResultSet |
getCharacteristics()
Returns some information characterizing or describing the current instance. |
protected StringBuffer |
getFurtherInformation()
This method is used in the subclasses to append further information to the XML representation. |
int |
getIndexOfMaximalComponentFor(Sequence s)
Returns the index i of the component with
P(i|s) |
String |
getInstanceName()
Should return a short instance name such as iMM(0), BN(2), ... |
double |
getLogPriorTerm()
Returns a value that is proportional to the log of the prior. |
protected double |
getLogPriorTermForComponentProbs()
This method computes the part of the prior that comes from the component probabilities. |
double |
getLogProbFor(int component,
Sequence s)
Returns the logarithmic probability for the sequence and the given component. |
double |
getLogProbFor(Sequence sequence,
int startpos,
int endpos)
Returns the logarithm of the probability of (a part of) the given sequence given the model. |
protected abstract double |
getLogProbUsingCurrentParameterSetFor(int component,
Sequence s,
int start,
int end)
Returns the logarithmic probability for the sequence and the given component using the current parameter set. |
double[] |
getLogScoreFor(DataSet data)
This method computes the logarithm of the scores of all sequences in the given data set. |
TrainableStatisticalModel |
getModel(int i)
Returns a deep copy of the i-th model. |
TrainableStatisticalModel[] |
getModels()
Returns a deep copy of the models. |
protected MultivariateRandomGenerator |
getMRG()
This method creates the multivariate random generator that will be used during initialization. |
protected MRGParams |
getMRGParams()
This method creates the parameters used in a multivariate random generator while initialization. |
String |
getNameOfAlgorithm()
Returns the name of the used algorithm. |
protected void |
getNewComponentProbs(double[] weights)
Estimates the weights of each component. |
protected void |
getNewParameters(int iteration,
double[][] seqWeights,
double[] w)
This method trains the internal models on the internal data set and the given weights. |
protected void |
getNewParametersForModel(int modelIndex,
int iteration,
int sampleIndex,
double[] seqWeights)
This method trains the internal model with index modelIndex
on the internal data set and the given weights. |
protected abstract double |
getNewWeights(double[] dataWeights,
double[] w,
double[][] seqweights)
Computes sequence weights and returns the score. |
int |
getNumberOfComponents()
Returns the number of components the are modeled by this AbstractMixtureTrainSM. |
NumericalResultSet |
getNumericalCharacteristics()
Returns the subset of numerical values that are also returned by SequenceScore.getCharacteristics(). |
double |
getScoreForBestRun()
Returns the value of the optimized function from the best run of the last training. |
double[] |
getWeights()
This method returns a deep copy of the weights for each component. |
protected void |
initModelForSampling(int starts)
This method initializes the model for the sampling. |
protected void |
initWithPrior(double[] w)
This method sets the initial weights before counting the usage of each component. |
boolean |
isInitialized()
This method can be used to determine whether the instance is initialized. |
protected boolean |
isInSamplingMode()
This method returns true if the object is currently used in
a sampling, otherwise false. |
double |
iterate(DataSet data,
double[] dataWeights,
MultivariateRandomGenerator m,
MRGParams[] params)
This method runs the train algorithm for the current model. |
protected double |
iterate(int start,
double[] dataWeights,
MultivariateRandomGenerator m,
MRGParams[] params)
This method runs the train algorithm for the current model and the internal data set. |
static int |
max(double[] w,
int start,
int end)
This method returns the index of a maximal entry in the array w between index start and end. |
protected double |
modifyWeights(double[] w)
This method modifies the computed weights for one sequence and returns the score. |
protected boolean |
parseNextParameterSet()
This method allows the user to parse the next set of parameters (from a file). |
protected boolean |
parseParameterSet(int sampling,
int burnInIteration)
This method allows the user to parse the set of parameters with index burnInIteration of a specific sampling (from a
file). |
protected void |
samplingStopped()
This method is the opposite of the method initModelForSampling(int). |
void |
setAlpha(double alpha)
Sets the parameter of the Dirichlet distribution which is used when you invoke train to init the gammas. |
void |
setOutputStream(OutputStream o)
Sets the OutputStream that is used e.g. |
protected abstract void |
setTrainData(DataSet data)
This method is invoked by the train-method and sets for a
given data set the data set that should be used for train. |
protected void |
setWeights(double... weights)
Sets the weights of each component. |
protected void |
swap()
This method swaps the current component models with the alternative model. |
StringBuffer |
toXML()
This method returns an XML representation as StringBuffer of an
instance of the implementing class. |
void |
train(DataSet data,
double[] dataWeights)
Trains the TrainableStatisticalModel object given the data as DataSet using
the specified weights. |
| Methods inherited from class de.jstacs.sequenceScores.statisticalModels.trainable.AbstractTrainableStatisticalModel |
|---|
check, getAlphabetContainer, getLength, getLogProbFor, getLogProbFor, getLogScoreFor, getLogScoreFor, getLogScoreFor, getLogScoreFor, getMaximalMarkovOrder, toString, train |
| Methods inherited from class java.lang.Object |
|---|
equals, getClass, hashCode, notify, notifyAll, wait, wait, wait |
| Methods inherited from interface de.jstacs.sequenceScores.SequenceScore |
|---|
toString |
| Field Detail |
|---|
protected double[] weights
protected double[] logWeights
protected double[] componentHyperParams
protected TrainableStatisticalModel[] model
protected TrainableStatisticalModel[] alternativeModel
protected int starts
protected int dimension
protected double best
protected SafeOutputStream sostream
protected DataSet[] sample
StringBuffer when invoking toXML().
protected boolean estimateComponentProbs
protected boolean[] optimizeModel
protected AbstractMixtureTrainSM.Algorithm algorithm
protected boolean algorithmHasBeenRun
protected int initialIteration
protected int stationaryIteration
protected BurnInTest burnInTest
BurnInTest that is used to stop the sampling.
protected BufferedWriter filewriter
protected BufferedReader filereader
protected File[] file
protected int[] counter
protected int samplingIndex
protected double[] compProb
protected double[][] seqWeights
| Constructor Detail |
|---|
protected AbstractMixtureTrainSM(int length,
TrainableStatisticalModel[] models,
boolean[] optimizeModel,
int dimension,
int starts,
boolean estimateComponentProbs,
double[] componentHyperParams,
double[] weights,
AbstractMixtureTrainSM.Algorithm algorithm,
double alpha,
TerminationCondition tc,
AbstractMixtureTrainSM.Parameterization parametrization,
int initialIteration,
int stationaryIteration,
BurnInTest burnInTest)
throws CloneNotSupportedException,
IllegalArgumentException,
WrongAlphabetException
AbstractMixtureTrainSM. This constructor can be used
for any algorithm since it takes all necessary values as parameters.
length - the length used in this modelmodels - the single models building the AbstractMixtureTrainSM,
if the model is trained using AbstractMixtureTrainSM.Algorithm.GIBBS_SAMPLING
the models that will be adjusted have to implement
SamplingComponentoptimizeModel - an array of switches to determine whether a model should be
optimized or notdimension - the number of componentsstarts - the number of times the algorithm will be started in the
train-method, at least 1estimateComponentProbs - the switch for estimating the component probabilities in the
algorithm or to hold them fixed; if the component parameters
are fixed, the values of weights will be used,
otherwise the componentHyperParams will be
incorporated in the adjustmentcomponentHyperParams - the hyperparameters for the component assignment prior
estimateComponentProbs == true
null or has to have
length dimension
null or an array with all values zero (0)
then ML
parameterization
weights - null or the weights for the components (then
weights.length == dimension)algorithm - either AbstractMixtureTrainSM.Algorithm.EM or
AbstractMixtureTrainSM.Algorithm.GIBBS_SAMPLINGalpha - only for AbstractMixtureTrainSM.Algorithm.EMtrain to initialize the
gammas. It is recommended to use alpha = 1
(uniform distribution on a simplex).tc - only for AbstractMixtureTrainSM.Algorithm.EMTerminationCondition for stopping the EM-algorithm,
tc has to return true from TerminationCondition.isSimple()parametrization - only for AbstractMixtureTrainSM.Algorithm.EMAbstractMixtureTrainSM.Parameterization.THETA or
AbstractMixtureTrainSM.Parameterization.LAMBDA
AbstractMixtureTrainSM.Parameterization.LAMBDA
initialIteration - only for AbstractMixtureTrainSM.Algorithm.GIBBS_SAMPLINGstationaryIteration/starts)stationaryIteration - only for AbstractMixtureTrainSM.Algorithm.GIBBS_SAMPLINGburnInTest - only for AbstractMixtureTrainSM.Algorithm.GIBBS_SAMPLINGIllegalArgumentException - if
length dimension < 1 weights != null && weights.length != dimension
weights != null and it exists an i
where weights[i] < 0 starts
< 1 componentHyperParams are not
correct WrongAlphabetException - if not all models work on the same alphabet
CloneNotSupportedException - if the models can not be cloned
protected AbstractMixtureTrainSM(StringBuffer xml)
throws NonParsableException
Storable.
Creates a new AbstractMixtureTrainSM out of its XML representation.
xml - the XML representation of the model as StringBuffer
NonParsableException - if the StringBuffer can not be parsed| Method Detail |
|---|
public AbstractMixtureTrainSM clone()
throws CloneNotSupportedException
AbstractTrainableStatisticalModelObject's clone()-method.
clone in interface SequenceScoreclone in interface TrainableStatisticalModelclone in class AbstractTrainableStatisticalModelAbstractTrainableStatisticalModel
(the member-AlphabetContainer isn't deeply cloned since
it is assumed to be immutable). The type of the returned object
is defined by the class X directly inherited from
AbstractTrainableStatisticalModel. Hence X's
clone()-method should work as:Object o = (X)super.clone(); o defined by
X that are not of simple data-types like
int, double, ... have to be deeply
copied return o
CloneNotSupportedException - if something went wrong while cloningprotected MultivariateRandomGenerator getMRG()
getMRGParams()protected MRGParams getMRGParams()
getMRG()
public void train(DataSet data,
double[] dataWeights)
throws Exception
TrainableStatisticalModelTrainableStatisticalModel object given the data as DataSet using
the specified weights. The weight at position i belongs to the element at
position i. So the array weight should have the number of
sequences in the data set as dimension. (Optionally it is possible to use
weight == null if all weights have the value one.)train(data1); train(data2)
should be a fully trained model over data2 and not over
data1+data2. All parameters of the model were given by the
call of the constructor.
data - the given sequences as DataSetdataWeights - the weights of the elements, each weight should be
non-negative
Exception - if the training did not succeed (e.g. the dimension of
weights and the number of sequences in the
data set do not match)DataSet.getElementAt(int),
DataSet.ElementEnumeratorprotected void swap()
train-method.
protected abstract void setTrainData(DataSet data)
throws Exception
train-method and sets for a
given data set the data set that should be used for train.
data - the given data set of sequences
Exception - if something went wrongprotected double[][] createSeqWeightsArray()
public double iterate(DataSet data,
double[] dataWeights,
MultivariateRandomGenerator m,
MRGParams[] params)
throws Exception
data - the data set of sequencesdataWeights - the weights for each sequence or nullm - the random generator for initiating the algorithmparams - the parameters for the sequences
Exception - if something went wrongdoFirstIteration(DataSet, double[],
MultivariateRandomGenerator, MRGParams[]),
continueIterations(double[], double[][]),
continueIterations(double[], double[][], int,
int)
protected double iterate(int start,
double[] dataWeights,
MultivariateRandomGenerator m,
MRGParams[] params)
throws Exception
start - the index of the trainingdataWeights - the weights for each sequence or nullm - the random generator for initiating the algorithmparams - the parameters for the sequences
Exception - if something went wrongdoFirstIteration(DataSet, double[],
MultivariateRandomGenerator, MRGParams[]),
continueIterations(double[], double[][]),
continueIterations(double[], double[][], int,
int)
protected double[][] doFirstIteration(DataSet data,
double[] dataWeights)
throws Exception
data - the data set of sequencesdataWeights - null or the weights of each element of the data set
Exception - if something went wrong
protected double[][] doFirstIteration(DataSet data,
double[] dataWeights,
MultivariateRandomGenerator m,
MRGParams[] params)
throws Exception
data - the data set of sequencesdataWeights - null or the weights of each element of the data setm - the multivariate random generatorparams - the parameters for the multivariate random generator
Exception - if something went wrong
protected abstract double[][] doFirstIteration(double[] dataWeights,
MultivariateRandomGenerator m,
MRGParams[] params)
throws Exception
dataWeights - null or the weights of each element of the data setm - the multivariate random generatorparams - the parameters for the multivariate random generator
Exception - if something went wrong
protected double continueIterations(double[] dataWeights,
double[][] seqweights)
throws Exception
AbstractMixtureTrainSM. So in this case the models have to be
trained already. This method is useful for restarting the train algorithm
at a certain point. The algorithm will stop if the difference between the
optimized functions for two iterations is smaller than the specified
threshold.
dataWeights - null or the weights of each element of the
internal data set (last data set the AbstractMixtureTrainSM
was trained on)seqweights - null or an array for weighting the sequences, see
createSeqWeightsArray()
Exception - if something went wrong
protected double continueIterations(double[] dataWeights,
double[][] seqweights,
int iterations,
int start)
throws Exception
AbstractMixtureTrainSM. So in this case the models have to be
trained already. This method is useful for restarting the algorithm at a
certain point. The algorithm will stop after the number of iterations.
dataWeights - null or the weights of each element of the
internal sample (last sample the AbstractMixtureTrainSM
was trained on)seqweights - null or an array for weighting the sequences, see
createSeqWeightsArray()iterations - the number of iterations that should be donestart - the index of the run in a TrainableStatisticalModel.train(DataSet)-call
Exception - if something went wrong
protected void getNewParameters(int iteration,
double[][] seqWeights,
double[] w)
throws Exception
iteration - the number of times this method has been invokedseqWeights - the weights for each model and sequencew - the weights for the components
Exception - if the training of the internal models went wrong
protected void getNewParametersForModel(int modelIndex,
int iteration,
int sampleIndex,
double[] seqWeights)
throws Exception
modelIndex
on the internal data set and the given weights.
modelIndex - the index of the modeliteration - the number of times this method has been invoked for this
modelsampleIndex - the index of the internal data set that should be usedseqWeights - the weights for each sequence
Exception - if the training of the internal model went wrong
protected abstract double getNewWeights(double[] dataWeights,
double[] w,
double[][] seqweights)
throws Exception
dataWeights - the weights for the internal data set (should not be changed)w - the array for the statistic of the component parameters (shall
be filled)seqweights - an array containing for each component the weights for each
sequence (shall be filled)
Exception - if something went wrongprotected double modifyWeights(double[] w)
w - the weights
protected void initWithPrior(double[] w)
w - the array of weights
public double getLogProbFor(int component,
Sequence s)
throws Exception
component - the index of the components - the sequence
log P(s,component) = log P(s|component) + log P(component)
Exception - if the model was not trained yet or something else went wronggetNumberOfComponents()
protected abstract double getLogProbUsingCurrentParameterSetFor(int component,
Sequence s,
int start,
int end)
throws Exception
component - the index of the components - the sequencestart - the start position in the sequenceend - the end position in the sequence
log P(s,component) = log P(s|component) + log P(component)
Exception - if not trained yet or something else went wronggetNumberOfComponents()
public final double getLogProbFor(Sequence sequence,
int startpos,
int endpos)
throws Exception
StatisticalModelStatisticalModel.getLogProbFor(Sequence, int) by the fact, that the model could be
e.g. homogeneous and therefore the length of the sequences, whose
probability should be returned, is not fixed. Additionally, the end
position of the part of the given sequence is given and the probability
of the part from position startpos to endpos
(inclusive) should be returned.
length and the alphabets define the type of
data that can be modeled and therefore both has to be checked.
sequence - the given sequencestartpos - the start position within the given sequenceendpos - the last position to be taken into account
Exception - if the sequence could not be handled (e.g.
startpos > , endpos
> sequence.length, ...) by the model
NotTrainedException - if the model is not trained yet
public final double[] getLogScoreFor(DataSet data)
throws Exception
SequenceScoreSequenceScore.getLogScoreFor(Sequence).
getLogScoreFor in interface SequenceScoregetLogScoreFor in class AbstractTrainableStatisticalModeldata - the data set of sequences
Exception - if something went wrongSequenceScore.getLogScoreFor(Sequence)
public double getLogPriorTerm()
throws Exception
StatisticalModel
Exception - if something went wrongprotected final double getLogPriorTermForComponentProbs()
public final double getScoreForBestRun()
throws NotTrainedException,
OperationNotSupportedException
NotTrainedException - if the training algorithm has not been run
OperationNotSupportedException - if this method is used for an instance that does not use the
EMtrain(DataSet, double[]),
algorithmHasBeenRun()public String getInstanceName()
SequenceScore
public int getIndexOfMaximalComponentFor(Sequence s)
throws Exception
i of the component with
P(i|s) maximal. Therefore it computes
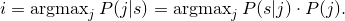
This method can be helpful for clustering.
- Parameters:
s - the sequence
- Returns:
- the index of the component
- Throws:
Exception - if the model was not trained yet or something else went wrong- See Also:
getLogProbFor(int, Sequence)
public final TrainableStatisticalModel[] getModels()
throws CloneNotSupportedException
AbstractTrainableStatisticalModels
CloneNotSupportedException - if at least one model can not be clonedgetModel(int)
public final TrainableStatisticalModel getModel(int i)
throws CloneNotSupportedException
i-th model.
i - the index
i-th model
CloneNotSupportedException - if at least one model can not be clonedgetModels()public String getNameOfAlgorithm()
public final int getNumberOfComponents()
AbstractMixtureTrainSM.
public ResultSet getCharacteristics()
throws Exception
SequenceScoreStorableResult.
getCharacteristics in interface SequenceScoregetCharacteristics in class AbstractTrainableStatisticalModelException - if some of the characteristics could not be definedStorableResult
public NumericalResultSet getNumericalCharacteristics()
throws Exception
SequenceScoreSequenceScore.getCharacteristics().
Exception - if some of the characteristics could not be definedpublic final double[] getWeights()
public boolean algorithmHasBeenRun()
true if the internal algorithm has been used to
determine the parameters of the modelpublic boolean isInitialized()
SequenceScoreSequenceScore.getLogScoreFor(Sequence).
true if the instance is initialized, false
otherwise
public final void setAlpha(double alpha)
throws IllegalArgumentException
train to init the gammas. It is recommended to use
alpha = 1 (uniform distribution on a simplex).
alpha - the parameter of the Dirichlet distribution with
alpha > 0
IllegalArgumentException - if alpha <= 0public final void setOutputStream(OutputStream o)
OutputStream that is used e.g. for writing information
while training. It is possible to set o=null, than nothing
will be written.
o - the OutputStream
protected void getNewComponentProbs(double[] weights)
throws Exception
weights - the array of weights, every element has to be non-negative and
the dimension has to be dimension
Exception - a weight is less than 0getNumberOfComponents()
protected void setWeights(double... weights)
throws IllegalArgumentException
weights - every element has to be non-negative, the sum of all weights
has to be 1 and the dimension of weights has to
be dimension
IllegalArgumentException - a weight is less than 0, the sum is not equal to 1 or the
dimension is incorrectgetNumberOfComponents()public StringBuffer toXML()
StorableStringBuffer of an
instance of the implementing class.
protected StringBuffer getFurtherInformation()
extractFurtherInformation(StringBuffer)
protected void fromXML(StringBuffer representation)
throws NonParsableException
AbstractTrainableStatisticalModelStringBuffer. It is the counter part of Storable.toXML().
fromXML in class AbstractTrainableStatisticalModelrepresentation - the XML representation of the model
NonParsableException - if the StringBuffer is not parsable or the
representation is conflictingAbstractTrainableStatisticalModel.AbstractTrainableStatisticalModel(StringBuffer)
protected void extractFurtherInformation(StringBuffer xml)
throws NonParsableException
xml - the XML representation
NonParsableException - if the XML representation is not parsablegetFurtherInformation()protected void checkModelsForGibbsSampling()
SamplingComponent.
protected void checkLength(int index,
int l)
l of the model with index
index is capable for the current instance. Otherwise an
IllegalArgumentException is thrown.
index - the index of the modell - the length of the model
IllegalArgumentException - if the model instance can not be used
public DataSet emitDataSet(int n,
int... lengths)
throws Exception
StatisticalModelDataSet object containing artificial
sequence(s).
emitDataSet( int n, int l ) should return a data set with
n sequences of length l.
emitDataSet( int n, int[] l ) should return a data set with
n sequences which have a sequence length corresponding to
the entry in the given array l.
emitDataSet( int n ) and
emitDataSet( int n, null ) should return a data set with
n sequences of length of the model (
SequenceScore.getLength()).
Exception.
emitDataSet in interface StatisticalModelemitDataSet in class AbstractTrainableStatisticalModeln - the number of sequences that should be contained in the
returned data setlengths - the length of the sequences for a homogeneous model; for an
inhomogeneous model this parameter should be null
or an array of size 0.
DataSet containing the artificial sequence(s)
Exception - if the emission did not succeed
NotTrainedException - if the model is not trained yetDataSet
protected abstract Sequence[] emitDataSetUsingCurrentParameterSet(int n,
int... lengths)
throws Exception
n - the number of sequences to be sampledlengths - the corresponding lengths
Exception - if it was impossible to sample the sequencesStatisticalModel.emitDataSet(int, int...)
protected boolean parseParameterSet(int sampling,
int burnInIteration)
throws Exception
burnInIteration of a specific sampling (from a
file).
sampling - the index of the samplingburnInIteration - the number of iterations that should be skipped
true if the parameter set could be parsed
Exception - if something went wrong while reading or parsing the
parameter set
protected boolean parseNextParameterSet()
throws Exception
true if the parameter set could be parsed
Exception - if something went wrong while reading or parsing the
parameter set
protected void initModelForSampling(int starts)
throws IOException
starts - the number of sampling starts
IOException - if the files could not be handled properly
protected void extendSampling(int sampling)
throws Exception
sampling - the index of the sampling
Exception - if the internal files could not be handled properly
protected void samplingStopped()
throws IOException
initModelForSampling(int). It can be used for closing any
streams of writer, ...
IOException - if the FileWriter could not be closed properlyprotected boolean isInSamplingMode()
true if the object is currently used in
a sampling, otherwise false.
true if the object is currently used in a sampling
protected void finalize()
throws Throwable
finalize in class ObjectThrowable
public static final int draw(double[] w,
int start)
w - an array containing probabilities starting at position
startstart - the start index
public static final int max(double[] w,
int start,
int end)
w between index start and end.
w - an arraystart - the start index (inclusive)end - the end index (exclusive)
|
||||||||||
| PREV CLASS NEXT CLASS | FRAMES NO FRAMES | |||||||||
| SUMMARY: NESTED | FIELD | CONSTR | METHOD | DETAIL: FIELD | CONSTR | METHOD | |||||||||