- mapDataSet(DataSet[]) - Method in class de.jstacs.classifiers.MappingClassifier
-
This method maps the given
DataSets to the internal classes.
- MappedDiscreteSequence - Class in de.jstacs.data.sequences
-
- MappedDiscreteSequence(AlphabetContainer, SequenceAnnotation[], DiscreteAlphabetMapping...) - Constructor for class de.jstacs.data.sequences.MappedDiscreteSequence
-
- MappedDiscreteSequence(SimpleDiscreteSequence, DiscreteAlphabetMapping...) - Constructor for class de.jstacs.data.sequences.MappedDiscreteSequence
-
- MappingClassifier - Class in de.jstacs.classifiers
-
This class allows the user to train the classifier on a given number of
classes and to evaluate the classifier on a smaller number of classes by
mapping classes together.
- MappingClassifier(AbstractScoreBasedClassifier, int...) - Constructor for class de.jstacs.classifiers.MappingClassifier
-
- MappingClassifier(StringBuffer) - Constructor for class de.jstacs.classifiers.MappingClassifier
-
The standard constructor for the interface
Storable.
- MappingDiffSM - Class in de.jstacs.sequenceScores.statisticalModels.differentiable
-
- MappingDiffSM(AlphabetContainer, DifferentiableStatisticalModel, DiscreteAlphabetMapping...) - Constructor for class de.jstacs.sequenceScores.statisticalModels.differentiable.MappingDiffSM
-
- MappingDiffSM(StringBuffer) - Constructor for class de.jstacs.sequenceScores.statisticalModels.differentiable.MappingDiffSM
-
- mapWeights(double[][]) - Method in class de.jstacs.classifiers.MappingClassifier
-
This method maps the given
Sequence weights to the internal classes.
- MarkovModelDiffSM - Class in de.jstacs.sequenceScores.statisticalModels.differentiable.directedGraphicalModels
-
- MarkovModelDiffSM(AlphabetContainer, int, double, boolean, int, DurationDiffSM) - Constructor for class de.jstacs.sequenceScores.statisticalModels.differentiable.directedGraphicalModels.MarkovModelDiffSM
-
This constructor creates an instance with an prior for the modeled length.
- MarkovModelDiffSM(AlphabetContainer, int, double, boolean, InhomogeneousMarkov) - Constructor for class de.jstacs.sequenceScores.statisticalModels.differentiable.directedGraphicalModels.MarkovModelDiffSM
-
This constructor creates an instance without any prior for the modeled length.
- MarkovModelDiffSM(AlphabetContainer, int, double, boolean, InhomogeneousMarkov, DurationDiffSM) - Constructor for class de.jstacs.sequenceScores.statisticalModels.differentiable.directedGraphicalModels.MarkovModelDiffSM
-
This constructor creates an instance with an prior for the modeled length.
- MarkovModelDiffSM(StringBuffer) - Constructor for class de.jstacs.sequenceScores.statisticalModels.differentiable.directedGraphicalModels.MarkovModelDiffSM
-
The standard constructor for the interface
Storable.
- MarkovRandomFieldDiffSM - Class in de.jstacs.sequenceScores.statisticalModels.differentiable
-
This class implements the scoring function for any MRF (Markov Random Field).
- MarkovRandomFieldDiffSM(AlphabetContainer, int, String) - Constructor for class de.jstacs.sequenceScores.statisticalModels.differentiable.MarkovRandomFieldDiffSM
-
- MarkovRandomFieldDiffSM(AlphabetContainer, int, double, String) - Constructor for class de.jstacs.sequenceScores.statisticalModels.differentiable.MarkovRandomFieldDiffSM
-
- MarkovRandomFieldDiffSM(StringBuffer) - Constructor for class de.jstacs.sequenceScores.statisticalModels.differentiable.MarkovRandomFieldDiffSM
-
This is the constructor for the interface
Storable.
- matches(int, Sequence) - Method in class de.jstacs.data.sequences.Sequence
-
This method allows to answer the question whether there is a
similar pattern find a match with a given maximal number of mismatches.
- MatrixCosts - Class in de.jstacs.algorithms.alignment.cost
-
Class for matrix costs, i.e., the cost of any match/mismatch is stored in
a matrix allowing a huge degree of freedom.
- MatrixCosts(double[][], double, double) - Constructor for class de.jstacs.algorithms.alignment.cost.MatrixCosts
-
Creates a new instance of
MatrixCosts where the costs
for mismatch and match are given in
matrix.
- MatrixCosts(double[][], double) - Constructor for class de.jstacs.algorithms.alignment.cost.MatrixCosts
-
Creates a new instance of
MatrixCosts where the costs
for mismatch and match are given in
matrix.
- MatrixCosts(StringBuffer) - Constructor for class de.jstacs.algorithms.alignment.cost.MatrixCosts
-
Restores
MatrixCosts object from its XML representation.
- matrixToString(double[][]) - Static method in class de.jstacs.utils.PFMComparator
-
Returns a string representation of the matrix, where each row of the matrix
is printed on one line and columns are separated by tabstops.
- max - Variable in class de.jstacs.sequenceScores.statisticalModels.differentiable.mixture.motif.DurationDiffSM
-
The maximal value.
- max(double[], int, int) - Static method in class de.jstacs.sequenceScores.statisticalModels.trainable.mixture.AbstractMixtureTrainSM
-
This method returns the index of a maximal entry in the array
w between index start and end.
- max - Variable in class de.jstacs.utils.DefaultProgressUpdater
-
The maximal number of steps.
- max(int, int) - Method in class de.jstacs.utils.DoubleList
-
This method computes the maximum of a part of the list.
- max(double...) - Static method in class de.jstacs.utils.ToolBox
-
This method returns the maximum of the elements of an array.
- max(int, int, double[]) - Static method in class de.jstacs.utils.ToolBox
-
This method returns the maximum of the elements of an array
between start and end.
- MaxHMMTrainingParameterSet - Class in de.jstacs.sequenceScores.statisticalModels.trainable.hmm.training
-
This class is the super class for any
HMMTrainingParameterSet that
is used for a maximizing training algorithm of a hidden Markov model.
- MaxHMMTrainingParameterSet() - Constructor for class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.training.MaxHMMTrainingParameterSet
-
This is the empty constructor that can be used to fill the parameters after creation.
- MaxHMMTrainingParameterSet(int, AbstractTerminationCondition) - Constructor for class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.training.MaxHMMTrainingParameterSet
-
This constructor can be used to create an instance with specified parameters.
- MaxHMMTrainingParameterSet(StringBuffer) - Constructor for class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.training.MaxHMMTrainingParameterSet
-
The standard constructor for the interface
Storable.
- MAXIMALBRANCHING - Static variable in class de.jstacs.algorithms.graphs.Chu_Liu_Edmonds
-
Compute the branching yielding the maximum sum of weights.
- maximalMarkovOrder - Variable in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.transitions.BasicHigherOrderTransition
-
The maximal Markov order of the transition.
- MaximumCorrelationCoefficient - Class in de.jstacs.classifiers.performanceMeasures
-
This class implements the maximum of the correlation coefficient
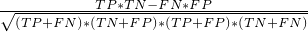
.
- MaximumCorrelationCoefficient() - Constructor for class de.jstacs.classifiers.performanceMeasures.MaximumCorrelationCoefficient
-
- MaximumCorrelationCoefficient(StringBuffer) - Constructor for class de.jstacs.classifiers.performanceMeasures.MaximumCorrelationCoefficient
-
The standard constructor for the interface
Storable.
- MaximumFMeasure - Class in de.jstacs.classifiers.performanceMeasures
-
Computes the maximum of the general F-measure given a positive real parameter
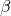
.
- MaximumFMeasure() - Constructor for class de.jstacs.classifiers.performanceMeasures.MaximumFMeasure
-
Constructs a new instance of the performance measure
MaximumFMeasure with empty parameters.
- MaximumFMeasure(double) - Constructor for class de.jstacs.classifiers.performanceMeasures.MaximumFMeasure
-
Constructs a new instance of the performance measure
MaximumFMeasure with given
beta.
- MaximumFMeasure(StringBuffer) - Constructor for class de.jstacs.classifiers.performanceMeasures.MaximumFMeasure
-
The standard constructor for the interface
Storable.
- MaximumNumericalTwoClassMeasure - Class in de.jstacs.classifiers.performanceMeasures
-
This class prepares everything for an easy implementation of a maximum of any numerical performance measure.
- MaximumNumericalTwoClassMeasure() - Constructor for class de.jstacs.classifiers.performanceMeasures.MaximumNumericalTwoClassMeasure
-
- MaximumNumericalTwoClassMeasure(StringBuffer) - Constructor for class de.jstacs.classifiers.performanceMeasures.MaximumNumericalTwoClassMeasure
-
The standard constructor for the interface
Storable.
- maxInDegree - Variable in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.transitions.BasicHigherOrderTransition
-
The maximal in-degree of any state.
- mean(int, int) - Method in class de.jstacs.utils.DoubleList
-
This method computes the mean of a part of the list.
- mean(int, int, double[]) - Static method in class de.jstacs.utils.ToolBox
-
This method returns the mean of the elements of an array
between start and end.
- MeanResultSet - Class in de.jstacs.results
-
- MeanResultSet(boolean, SimpleResult...) - Constructor for class de.jstacs.results.MeanResultSet
-
- MeanResultSet(SimpleResult...) - Constructor for class de.jstacs.results.MeanResultSet
-
- MeanResultSet() - Constructor for class de.jstacs.results.MeanResultSet
-
- MeanResultSet(StringBuffer) - Constructor for class de.jstacs.results.MeanResultSet
-
The standard constructor for the interface
Storable.
- MeanResultSet.AdditionImpossibleException - Exception in de.jstacs.results
-
Class for the exception that is thrown if two
MeanResultSets
should be added that do not match.
- MeanResultSet.InconsistentResultNumberException - Exception in de.jstacs.results
-
Class for the exception that is thrown if a
NumericalResultSet is
added to the
MeanResultSet that has a number of results which is
not equal to the number of results of the previously added results.
- MeanSmoothing(int) - Constructor for class de.jstacs.data.DinucleotideProperty.MeanSmoothing
-
- meanValue - Variable in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.states.emissions.continuous.PluginGaussianEmission
-
Initial mean value.
- Measure - Class in de.jstacs.sequenceScores.statisticalModels.differentiable.directedGraphicalModels.structureLearning.measures
-
Class for structure measures that derive an optimal structure with respect to
some criterion within a class of possible structures from data.
- Measure(StringBuffer) - Constructor for class de.jstacs.sequenceScores.statisticalModels.differentiable.directedGraphicalModels.structureLearning.measures.Measure
-
Creates a new
Measure from its XML-representation.
- Measure(Measure.MeasureParameterSet) - Constructor for class de.jstacs.sequenceScores.statisticalModels.differentiable.directedGraphicalModels.structureLearning.measures.Measure
-
- Measure() - Constructor for class de.jstacs.sequenceScores.statisticalModels.differentiable.directedGraphicalModels.structureLearning.measures.Measure
-
Default constructor.
- Measure.MeasureParameterSet - Class in de.jstacs.sequenceScores.statisticalModels.differentiable.directedGraphicalModels.structureLearning.measures
-
- MeasureParameterSet(Class<? extends Measure>) - Constructor for class de.jstacs.sequenceScores.statisticalModels.differentiable.directedGraphicalModels.structureLearning.measures.Measure.MeasureParameterSet
-
- MeasureParameterSet(StringBuffer) - Constructor for class de.jstacs.sequenceScores.statisticalModels.differentiable.directedGraphicalModels.structureLearning.measures.Measure.MeasureParameterSet
-
The standard constructor for the interface
Storable.
- median(int, int) - Method in class de.jstacs.utils.DoubleList
-
This method computes the median of a part of the list.
- median(double...) - Static method in class de.jstacs.utils.ToolBox
-
This method returns the median of the elements of array.
- median(int, int, double[]) - Static method in class de.jstacs.utils.ToolBox
-
This method returns the median of the elements of an array
between start and end.
- MedianSmoothing(int) - Constructor for class de.jstacs.data.DinucleotideProperty.MedianSmoothing
-
- MEM - Class in de.jstacs.sequenceScores.statisticalModels.trainable.discrete.inhomogeneous
-
This class represents a maximum entropy model.
- MEM(AbstractList<int[]>, int[], int[]) - Constructor for class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.inhomogeneous.MEM
-
The main constructor of a MEM.
- MEM(int[], int[], int[][]) - Constructor for class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.inhomogeneous.MEM
-
The main constructor of a MEM.
- MEM(StringBuffer) - Constructor for class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.inhomogeneous.MEM
-
The constructor for the Storable interface.
- MEManager - Class in de.jstacs.sequenceScores.statisticalModels.trainable.discrete.inhomogeneous
-
This class is the super class for all maximum entropy models
- MEManager(MEManagerParameterSet) - Constructor for class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.inhomogeneous.MEManager
-
- MEManager(StringBuffer) - Constructor for class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.inhomogeneous.MEManager
-
The standard constructor for the interface
Storable.
- MEManagerParameterSet - Class in de.jstacs.sequenceScores.statisticalModels.trainable.discrete.inhomogeneous.parameters
-
The ParameterSet for any MEManager.
- MEManagerParameterSet(StringBuffer) - Constructor for class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.inhomogeneous.parameters.MEManagerParameterSet
-
The constructor for the
Storable interface.
- MEManagerParameterSet(Class<? extends MEManager>) - Constructor for class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.inhomogeneous.parameters.MEManagerParameterSet
-
The constructor an empty constructor of extended class.
- MEManagerParameterSet(Class<? extends MEManager>, AlphabetContainer, int, double, String, ConstraintManager.Decomposition, boolean, byte, double) - Constructor for class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.inhomogeneous.parameters.MEManagerParameterSet
-
The fast constructor.
- MEMConstraint - Class in de.jstacs.sequenceScores.statisticalModels.trainable.discrete.inhomogeneous
-
This constraint can be used for any maximum entropy
model (MEM) application.
- MEMConstraint(int[], int[]) - Constructor for class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.inhomogeneous.MEMConstraint
-
- MEMConstraint(int[], int[], int[]) - Constructor for class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.inhomogeneous.MEMConstraint
-
- MEMConstraint(StringBuffer) - Constructor for class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.inhomogeneous.MEMConstraint
-
The standard constructor for the interface
Storable.
- MEMTools - Class in de.jstacs.sequenceScores.statisticalModels.trainable.discrete.inhomogeneous
-
- MEMTools() - Constructor for class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.inhomogeneous.MEMTools
-
- MEMTools.DualFunction - Class in de.jstacs.sequenceScores.statisticalModels.trainable.discrete.inhomogeneous
-
The dual function to the constraint problem of learning MEM's.
- merge(Hashtable<Sequence, BitSet[]>, int, boolean) - Static method in class de.jstacs.motifDiscovery.KMereStatistic
-
This method allows to merge the statistics of k-mers by allowing mismatches.
- min - Variable in class de.jstacs.sequenceScores.statisticalModels.differentiable.mixture.motif.DurationDiffSM
-
The minimal value.
- min(int, int) - Method in class de.jstacs.utils.DoubleList
-
This method computes the minimum of a part of the list.
- min(double...) - Static method in class de.jstacs.utils.ToolBox
-
This method returns the minimum of the elements of an array.
- min(int, int, double[]) - Static method in class de.jstacs.utils.ToolBox
-
This method returns the minimum of the elements of an array
between start and end.
- MINIMALBRANCHING - Static variable in class de.jstacs.algorithms.graphs.Chu_Liu_Edmonds
-
Compute the branching yielding the minimum sum of weights.
- minStepSize - Static variable in class de.jstacs.classifiers.performanceMeasures.PRCurve
-
The minimum step size between supporting points
- MixtureDiffSM - Class in de.jstacs.sequenceScores.statisticalModels.differentiable.mixture
-
This class implements a real mixture model.
- MixtureDiffSM(int, boolean, DifferentiableStatisticalModel...) - Constructor for class de.jstacs.sequenceScores.statisticalModels.differentiable.mixture.MixtureDiffSM
-
- MixtureDiffSM(StringBuffer) - Constructor for class de.jstacs.sequenceScores.statisticalModels.differentiable.mixture.MixtureDiffSM
-
This is the constructor for the interface
Storable.
- MixtureDurationDiffSM - Class in de.jstacs.sequenceScores.statisticalModels.differentiable.mixture.motif
-
- MixtureDurationDiffSM(int, DurationDiffSM...) - Constructor for class de.jstacs.sequenceScores.statisticalModels.differentiable.mixture.motif.MixtureDurationDiffSM
-
- MixtureDurationDiffSM(StringBuffer) - Constructor for class de.jstacs.sequenceScores.statisticalModels.differentiable.mixture.motif.MixtureDurationDiffSM
-
- MixtureEmission - Class in de.jstacs.sequenceScores.statisticalModels.trainable.hmm.states.emissions
-
This class implements a mixture of
Emissions.
- MixtureEmission(Emission[], double[]) - Constructor for class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.states.emissions.MixtureEmission
-
- MixtureEmission(StringBuffer) - Constructor for class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.states.emissions.MixtureEmission
-
The standard constructor for the interface
Storable.
- MixtureTrainSM - Class in de.jstacs.sequenceScores.statisticalModels.trainable.mixture
-
- MixtureTrainSM(int, TrainableStatisticalModel[], int, boolean, double[], double[], AbstractMixtureTrainSM.Algorithm, double, TerminationCondition, AbstractMixtureTrainSM.Parameterization, int, int, BurnInTest) - Constructor for class de.jstacs.sequenceScores.statisticalModels.trainable.mixture.MixtureTrainSM
-
- MixtureTrainSM(int, TrainableStatisticalModel[], int, double[], double, TerminationCondition, AbstractMixtureTrainSM.Parameterization) - Constructor for class de.jstacs.sequenceScores.statisticalModels.trainable.mixture.MixtureTrainSM
-
Creates an instance using EM and estimating the component probabilities.
- MixtureTrainSM(int, TrainableStatisticalModel[], double[], int, double, TerminationCondition, AbstractMixtureTrainSM.Parameterization) - Constructor for class de.jstacs.sequenceScores.statisticalModels.trainable.mixture.MixtureTrainSM
-
Creates an instance using EM and fixed component probabilities.
- MixtureTrainSM(int, TrainableStatisticalModel[], int, double[], int, int, BurnInTest) - Constructor for class de.jstacs.sequenceScores.statisticalModels.trainable.mixture.MixtureTrainSM
-
Creates an instance using Gibbs Sampling and sampling the component
probabilities.
- MixtureTrainSM(int, TrainableStatisticalModel[], double[], int, int, int, BurnInTest) - Constructor for class de.jstacs.sequenceScores.statisticalModels.trainable.mixture.MixtureTrainSM
-
Creates an instance using Gibbs Sampling and fixed component
probabilities.
- MixtureTrainSM(StringBuffer) - Constructor for class de.jstacs.sequenceScores.statisticalModels.trainable.mixture.MixtureTrainSM
-
The constructor for the interface
Storable.
- model - Variable in class de.jstacs.sequenceScores.statisticalModels.trainable.mixture.AbstractMixtureTrainSM
-
The model for the sequences.
- models - Variable in class de.jstacs.classifiers.trainSMBased.TrainSMBasedClassifier
-
- models - Variable in class de.jstacs.sequenceScores.statisticalModels.trainable.CompositeTrainSM
-
The models for the components
- modify(int, int) - Method in interface de.jstacs.motifDiscovery.Mutable
-
Manually modifies the model.
- modify(int, int) - Method in class de.jstacs.sequenceScores.statisticalModels.differentiable.directedGraphicalModels.MarkovModelDiffSM
-
- modify(int, int) - Method in class de.jstacs.sequenceScores.statisticalModels.differentiable.localMixture.LimitedSparseLocalInhomogeneousMixtureDiffSM_higherOrder
-
- modify(int, int) - Method in class de.jstacs.sequenceScores.statisticalModels.differentiable.MappingDiffSM
-
- modify(int, int) - Method in class de.jstacs.sequenceScores.statisticalModels.differentiable.MarkovRandomFieldDiffSM
-
- modify(int) - Method in class de.jstacs.sequenceScores.statisticalModels.differentiable.mixture.motif.DurationDiffSM
-
- modify(int) - Method in class de.jstacs.sequenceScores.statisticalModels.differentiable.mixture.motif.MixtureDurationDiffSM
-
- modify(int) - Method in class de.jstacs.sequenceScores.statisticalModels.differentiable.mixture.motif.SkewNormalLikeDurationDiffSM
-
- modify(int) - Method in class de.jstacs.sequenceScores.statisticalModels.differentiable.mixture.motif.UniformDurationDiffSM
-
- modify(int, int) - Method in class de.jstacs.sequenceScores.statisticalModels.differentiable.mixture.StrandDiffSM
-
- modify(int, int) - Method in class de.jstacs.sequenceScores.statisticalModels.differentiable.NormalizedDiffSM
-
- modify(double[], double[], int, int) - Method in class de.jstacs.sequenceScores.statisticalModels.trainable.mixture.motif.ZOOPSTrainSM
-
This method modifies the computed weights for one sequence and returns
the score.
- modifyFunctionValue(double) - Method in class de.jstacs.classifiers.differentiableSequenceScoreBased.sampling.SamplingGenDisMixClassifier
-
- modifyFunctionValue(double) - Method in class de.jstacs.classifiers.differentiableSequenceScoreBased.sampling.SamplingScoreBasedClassifier
-
- modifyMotif(int, int, int) - Method in interface de.jstacs.motifDiscovery.MutableMotifDiscoverer
-
Manually modifies the motif model with index motifIndex.
- modifyMotif(int, int, int) - Method in class de.jstacs.sequenceScores.statisticalModels.differentiable.IndependentProductDiffSM
-
- modifyMotif(int, int, int) - Method in class de.jstacs.sequenceScores.statisticalModels.differentiable.MappingDiffSM
-
- modifyMotif(int, int, int) - Method in class de.jstacs.sequenceScores.statisticalModels.differentiable.mixture.MixtureDiffSM
-
- modifyMotif(int, int, int) - Method in class de.jstacs.sequenceScores.statisticalModels.differentiable.mixture.motif.ExtendedZOOPSDiffSM
-
- modifyWeights(double[]) - Method in class de.jstacs.sequenceScores.statisticalModels.trainable.mixture.AbstractMixtureTrainSM
-
This method modifies the computed weights for one sequence and returns
the score.
- MotifAnnotation - Class in de.jstacs.data.sequences.annotation
-
- MotifAnnotation(String, int, int, StrandedLocatedSequenceAnnotationWithLength.Strand, Result...) - Constructor for class de.jstacs.data.sequences.annotation.MotifAnnotation
-
Creates a new
MotifAnnotation of type
type with
identifier
identifier and additional annotation (that does
not fit the
SequenceAnnotation definitions) given as an array of
Results
additionalAnnotation.
- MotifAnnotation(StringBuffer) - Constructor for class de.jstacs.data.sequences.annotation.MotifAnnotation
-
The standard constructor for the interface
Storable.
- MotifAnnotationParser - Class in de.jstacs.data.sequences.annotation
-
- MotifAnnotationParser() - Constructor for class de.jstacs.data.sequences.annotation.MotifAnnotationParser
-
- MotifAnnotationParser(String, String) - Constructor for class de.jstacs.data.sequences.annotation.MotifAnnotationParser
-
- MotifDiscoverer - Interface in de.jstacs.motifDiscovery
-
This is the interface that any tool for de-novo motif discovery should
implement.
- MotifDiscoverer.KindOfProfile - Enum in de.jstacs.motifDiscovery
-
This enum can be used to determine which kind of profile
should be returned.
- MotifDiscovererToolBox - Class in de.jstacs.motifDiscovery
-
- MotifDiscovererToolBox() - Constructor for class de.jstacs.motifDiscovery.MotifDiscovererToolBox
-
- MotifDiscoveryAssessment - Class in de.jstacs.motifDiscovery
-
This class enables the user to assess the prediction of motif occurrences
- MotifDiscoveryAssessment() - Constructor for class de.jstacs.motifDiscovery.MotifDiscoveryAssessment
-
- motifLength - Variable in class de.jstacs.sequenceScores.statisticalModels.trainable.mixture.motif.positionprior.PositionPrior
-
The length of the motif.
- MRGParams - Class in de.jstacs.utils.random
-
The super container for parameter of multivariate random generators.
- MRGParams() - Constructor for class de.jstacs.utils.random.MRGParams
-
- MSPClassifier - Class in de.jstacs.classifiers.differentiableSequenceScoreBased.msp
-
This class implements a classifier that allows the training via MCL or MSP principle.
- MSPClassifier(GenDisMixClassifierParameterSet, DifferentiableSequenceScore...) - Constructor for class de.jstacs.classifiers.differentiableSequenceScoreBased.msp.MSPClassifier
-
This convenience constructor creates an
MSPClassifier that used MCL principle for training.
- MSPClassifier(GenDisMixClassifierParameterSet, LogPrior, DifferentiableSequenceScore...) - Constructor for class de.jstacs.classifiers.differentiableSequenceScoreBased.msp.MSPClassifier
-
- MSPClassifier(GenDisMixClassifierParameterSet, LogPrior, double, DifferentiableSequenceScore...) - Constructor for class de.jstacs.classifiers.differentiableSequenceScoreBased.msp.MSPClassifier
-
- MSPClassifier(StringBuffer) - Constructor for class de.jstacs.classifiers.differentiableSequenceScoreBased.msp.MSPClassifier
-
- MST - Class in de.jstacs.algorithms.graphs
-
This class enables you to compute the maximal spanning forest for an
undirected, weighted graph.
- MST() - Constructor for class de.jstacs.algorithms.graphs.MST
-
- MultiDimensionalArbitrarySequence - Class in de.jstacs.data.sequences
-
This class is for multidimensional arbitrary sequences.
- MultiDimensionalArbitrarySequence(SequenceAnnotation[], ArbitrarySequence...) - Constructor for class de.jstacs.data.sequences.MultiDimensionalArbitrarySequence
-
- MultiDimensionalDiscreteSequence - Class in de.jstacs.data.sequences
-
This class is for multidimensional discrete sequences that can be used, for instance, for phylogenetic footprinting.
- MultiDimensionalDiscreteSequence(SequenceAnnotation[], SimpleDiscreteSequence...) - Constructor for class de.jstacs.data.sequences.MultiDimensionalDiscreteSequence
-
- MultiDimensionalSequence<T> - Class in de.jstacs.data.sequences
-
This class is for multidimensional sequences that can be used, for instance, for phylogenetic footprinting.
- MultiDimensionalSequence(SequenceAnnotation[], Sequence...) - Constructor for class de.jstacs.data.sequences.MultiDimensionalSequence
-
- MultiDimensionalSequenceWrapperDiffSS - Class in de.jstacs.sequenceScores.differentiable
-
This class implements a simple wrapper for multidimensional sequences.
- MultiDimensionalSequenceWrapperDiffSS(DifferentiableSequenceScore) - Constructor for class de.jstacs.sequenceScores.differentiable.MultiDimensionalSequenceWrapperDiffSS
-
The main constructor.
- MultiDimensionalSequenceWrapperDiffSS(StringBuffer) - Constructor for class de.jstacs.sequenceScores.differentiable.MultiDimensionalSequenceWrapperDiffSS
-
- MultilineSimpleParameter - Class in de.jstacs.tools.ui.galaxy
-
- MultilineSimpleParameter(String, String, boolean, Object) - Constructor for class de.jstacs.tools.ui.galaxy.MultilineSimpleParameter
-
- MultilineSimpleParameter(String, String, boolean, ParameterValidator, Object) - Constructor for class de.jstacs.tools.ui.galaxy.MultilineSimpleParameter
-
- MultilineSimpleParameter(String, String, boolean, ParameterValidator) - Constructor for class de.jstacs.tools.ui.galaxy.MultilineSimpleParameter
-
- MultilineSimpleParameter(String, String, boolean) - Constructor for class de.jstacs.tools.ui.galaxy.MultilineSimpleParameter
-
- MultilineSimpleParameter(StringBuffer) - Constructor for class de.jstacs.tools.ui.galaxy.MultilineSimpleParameter
-
The standard constructor for the interface
Storable.
- MultipleIterationsCondition - Class in de.jstacs.algorithms.optimization.termination
-
- MultipleIterationsCondition(int, AbstractTerminationCondition) - Constructor for class de.jstacs.algorithms.optimization.termination.MultipleIterationsCondition
-
This constructor creates an instance that stops the optimization if provided termination condition
fails a contiguously a specified number of times.
- MultipleIterationsCondition(MultipleIterationsCondition.MultipleIterationsConditionParameterSet) - Constructor for class de.jstacs.algorithms.optimization.termination.MultipleIterationsCondition
-
This is the main constructor creating an instance from a given parameter set.
- MultipleIterationsCondition(StringBuffer) - Constructor for class de.jstacs.algorithms.optimization.termination.MultipleIterationsCondition
-
The standard constructor for the interface
Storable.
- MultipleIterationsCondition.MultipleIterationsConditionParameterSet - Class in de.jstacs.algorithms.optimization.termination
-
- MultipleIterationsConditionParameterSet() - Constructor for class de.jstacs.algorithms.optimization.termination.MultipleIterationsCondition.MultipleIterationsConditionParameterSet
-
This constructor creates an empty parameter set.
- MultipleIterationsConditionParameterSet(StringBuffer) - Constructor for class de.jstacs.algorithms.optimization.termination.MultipleIterationsCondition.MultipleIterationsConditionParameterSet
-
The standard constructor for the interface
Storable.
- MultipleIterationsConditionParameterSet(int, AbstractTerminationCondition) - Constructor for class de.jstacs.algorithms.optimization.termination.MultipleIterationsCondition.MultipleIterationsConditionParameterSet
-
This constructor creates a filled instance of the parameter set.
- multiply(int, int, double) - Method in class de.jstacs.utils.DoubleList
-
Multiplies all values in the list from index start to
end with the value factor.
- multiplyExpLambdaWith(int, double) - Method in class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.inhomogeneous.MEMConstraint
-
Multiplies the exponential value of
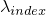
with the factor
val:
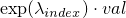
.
- MultiSelectionParameter - Class in de.jstacs.parameters
-
Class for a
Parameter that provides a collection of possible values.
- MultiSelectionParameter(DataType, String[], Object[], String, String, boolean) - Constructor for class de.jstacs.parameters.MultiSelectionParameter
-
- MultiSelectionParameter(DataType, String[], Object[], String[], String, String, boolean) - Constructor for class de.jstacs.parameters.MultiSelectionParameter
-
- MultiSelectionParameter(String, String, boolean, ParameterSet...) - Constructor for class de.jstacs.parameters.MultiSelectionParameter
-
- MultiSelectionParameter(String, String, boolean, Class<? extends ParameterSet>...) - Constructor for class de.jstacs.parameters.MultiSelectionParameter
-
- MultiSelectionParameter(StringBuffer) - Constructor for class de.jstacs.parameters.MultiSelectionParameter
-
The standard constructor for the interface
Storable.
- MultiThreadedFunction - Interface in de.jstacs.algorithms.optimization
-
This interface defines methods for functions that are multi-threaded.
- MultiThreadedTrainingParameterSet - Class in de.jstacs.sequenceScores.statisticalModels.trainable.hmm.training
-
This class is the super class for any
MaxHMMTrainingParameterSet that
is used for a multi-threaded maximizing training algorithm of a hidden Markov model.
- MultiThreadedTrainingParameterSet() - Constructor for class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.training.MultiThreadedTrainingParameterSet
-
This is the empty constructor that can be used to fill the parameters after creation.
- MultiThreadedTrainingParameterSet(int, AbstractTerminationCondition, int) - Constructor for class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.training.MultiThreadedTrainingParameterSet
-
This constructor can be used to create an instance with specified parameters.
- MultiThreadedTrainingParameterSet(StringBuffer) - Constructor for class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.training.MultiThreadedTrainingParameterSet
-
The standard constructor for the interface
Storable.
- MultivariateGaussianEmission - Class in de.jstacs.sequenceScores.statisticalModels.trainable.hmm.states.emissions.continuous
-
Multivariate Gaussian emission density for a Hidden Markov Model.
- MultivariateGaussianEmission(double[], double[], double[][], double, double[], double, double[][]) - Constructor for class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.states.emissions.continuous.MultivariateGaussianEmission
-
Creates a Multivariate Gaussian emission density.
- MultivariateGaussianEmission(StringBuffer) - Constructor for class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.states.emissions.continuous.MultivariateGaussianEmission
-
- MultivariateRandomGenerator - Class in de.jstacs.utils.random
-
This class is the abstract super class for any multivariate random generator
(MRG).
- MultivariateRandomGenerator() - Constructor for class de.jstacs.utils.random.MultivariateRandomGenerator
-
- mus - Variable in class de.jstacs.classifiers.differentiableSequenceScoreBased.logPrior.SeparateGaussianLogPrior
-
The means
- Mutable - Interface in de.jstacs.motifDiscovery
-
This interface allows to modify a motif model.
- MutableMotifDiscoverer - Interface in de.jstacs.motifDiscovery
-
This is the interface that any tool for de-novo motif discovery should implement that allows any modify-operations like shift, shrink and expand.
- MutableMotifDiscovererToolbox - Class in de.jstacs.motifDiscovery
-
- MutableMotifDiscovererToolbox() - Constructor for class de.jstacs.motifDiscovery.MutableMotifDiscovererToolbox
-
- MutableMotifDiscovererToolbox.InitMethodForDiffSM - Enum in de.jstacs.motifDiscovery
-
- myAbstractClassifier - Variable in class de.jstacs.classifiers.assessment.ClassifierAssessment
-
This array contains the internal used classifiers.
- myModel - Variable in class de.jstacs.classifiers.assessment.ClassifierAssessment
-
This array contains for each class the internal used models.
- myTempMeanResultSets - Variable in class de.jstacs.classifiers.assessment.ClassifierAssessment
-
The temporary result set.
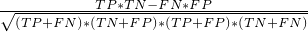 .
.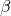 .
.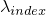 with the factor
with the factor 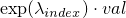 .
.