- d - Variable in class de.jstacs.algorithms.alignment.Alignment
-
The matrices holding the edit distances
- DAG - Class in de.jstacs.algorithms.graphs
-
This is the main class of the graph library.
- DAG() - Constructor for class de.jstacs.algorithms.graphs.DAG
-
- DAGTrainSM - Class in de.jstacs.sequenceScores.statisticalModels.trainable.discrete.inhomogeneous
-
The abstract class for
directed
acyclic
graphical models
(
DAGTrainSM).
- DAGTrainSM(IDGTrainSMParameterSet) - Constructor for class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.inhomogeneous.DAGTrainSM
-
This is the main constructor.
- DAGTrainSM(StringBuffer) - Constructor for class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.inhomogeneous.DAGTrainSM
-
The standard constructor for the interface
Storable.
- data - Variable in class de.jstacs.classifiers.differentiableSequenceScoreBased.AbstractOptimizableFunction
-
The data that is used to evaluate this function.
- DataColumnParameter - Class in de.jstacs.tools
-
SimpleParameter that represents a data column parameter in Galaxy and JstacsFX.
- DataColumnParameter(String, String, String, boolean, Integer) - Constructor for class de.jstacs.tools.DataColumnParameter
-
- DataColumnParameter(String, String, String, boolean, ParameterValidator, Integer) - Constructor for class de.jstacs.tools.DataColumnParameter
-
- DataColumnParameter(String, String, String, boolean, ParameterValidator) - Constructor for class de.jstacs.tools.DataColumnParameter
-
- DataColumnParameter(String, String, String, boolean) - Constructor for class de.jstacs.tools.DataColumnParameter
-
- DataColumnParameter(StringBuffer) - Constructor for class de.jstacs.tools.DataColumnParameter
-
The standard constructor for the interface
Storable.
- DataSet - Class in de.jstacs.data
-
This is the class for any data set of
Sequences.
- DataSet(AlphabetContainer, AbstractStringExtractor) - Constructor for class de.jstacs.data.DataSet
-
- DataSet(AlphabetContainer, AbstractStringExtractor, int) - Constructor for class de.jstacs.data.DataSet
-
- DataSet(AlphabetContainer, AbstractStringExtractor, String) - Constructor for class de.jstacs.data.DataSet
-
- DataSet(AlphabetContainer, AbstractStringExtractor, String, int) - Constructor for class de.jstacs.data.DataSet
-
- DataSet(AlphabetContainer, AbstractStringExtractor, String, int, double) - Constructor for class de.jstacs.data.DataSet
-
- DataSet(DataSet, int) - Constructor for class de.jstacs.data.DataSet
-
Creates a new
DataSet from a given
DataSet and a given
length
subsequenceLength.
This constructor enables you to use subsequences of the elements of a
DataSet.
- DataSet(String, Sequence...) - Constructor for class de.jstacs.data.DataSet
-
- DataSet(String, Collection<Sequence>) - Constructor for class de.jstacs.data.DataSet
-
- DataSet.ElementEnumerator - Class in de.jstacs.data
-
This class can be used to have a fast sequential access to a
DataSet.
- DataSet.PartitionMethod - Enum in de.jstacs.data
-
This
enum defines different partition methods for a
DataSet.
- DataSet.WeightedDataSetFactory - Class in de.jstacs.data
-
This class enables you to eliminate
Sequences that occur more
than once in one or more
DataSets.
- DataSet.WeightedDataSetFactory.SortOperation - Enum in de.jstacs.data
-
- DataSetKMerEnumerator - Class in de.jstacs.data
-
- DataSetKMerEnumerator(DataSet, int, boolean) - Constructor for class de.jstacs.data.DataSetKMerEnumerator
-
Constructs a new DataSetKMerEnumerator from a
DataSet data by extracting all k-mers.
- DataSetResult - Class in de.jstacs.results
-
- DataSetResult(String, String, DataSet) - Constructor for class de.jstacs.results.DataSetResult
-
- DataSetResult(StringBuffer) - Constructor for class de.jstacs.results.DataSetResult
-
The standard constructor for the interface
Storable.
- DataSetResultSaver - Class in de.jstacs.results.savers
-
- dataSetToSequenceIterator(DataSet, boolean, boolean) - Static method in class de.jstacs.data.bioJava.BioJavaAdapter
-
- datatype - Variable in class de.jstacs.AnnotatedEntity
-
The data type of the entity.
- DataType - Enum in de.jstacs
-
This
enum defines a number of data types that can be used for
Parameters and
Result
s.
- DatatypeNotValidException(String) - Constructor for exception de.jstacs.parameters.SimpleParameter.DatatypeNotValidException
-
- dataTypeToGalaxy() - Method in class de.jstacs.parameters.SimpleParameter
-
Returns the Galaxy identifier for the
DataType of this parameter
- dataTypeToGalaxy() - Method in class de.jstacs.tools.DataColumnParameter
-
- DateFileFilter - Class in de.jstacs.io
-
This class implements a
FileFilter that accepts
Files that were modified after the date that is given in the constructor.
- DateFileFilter(int, int, int, int, int, int) - Constructor for class de.jstacs.io.DateFileFilter
-
Creates an instance that accepts
Files that were modified after the given year, month, ...
- DateFileFilter(Date) - Constructor for class de.jstacs.io.DateFileFilter
-
Creates an instance that accepts
Files that were modified after
d.
- de.jstacs - package de.jstacs
-
This package is the root package for the most and important packages.
- de.jstacs.algorithms.alignment - package de.jstacs.algorithms.alignment
-
Provides classes for alignments.
- de.jstacs.algorithms.alignment.cost - package de.jstacs.algorithms.alignment.cost
-
Provides classes for cost functions used in alignments.
- de.jstacs.algorithms.graphs - package de.jstacs.algorithms.graphs
-
Provides classes for algorithms on graphs.
- de.jstacs.algorithms.graphs.tensor - package de.jstacs.algorithms.graphs.tensor
-
Provides classes to represent symmetric and asymmetric tensors in graphs.
- de.jstacs.algorithms.optimization - package de.jstacs.algorithms.optimization
-
Provides classes for different types of algorithms that are not directly linked to the modelling components of Jstacs: Algorithms on graphs, algorithms for numerical optimization, and a basic alignment algorithm.
- de.jstacs.algorithms.optimization.termination - package de.jstacs.algorithms.optimization.termination
-
Provides classes for termination conditions that can be used in algorithms.
- de.jstacs.classifiers - package de.jstacs.classifiers
-
This package provides the framework for any classifier.
- de.jstacs.classifiers.assessment - package de.jstacs.classifiers.assessment
-
This package allows to assess classifiers.
It contains the class ClassifierAssessment that
is used as a super-class of all implemented methodologies of
an assessment to assess classifiers.
- de.jstacs.classifiers.differentiableSequenceScoreBased - package de.jstacs.classifiers.differentiableSequenceScoreBased
-
Provides the classes for
Classifiers that are based on
SequenceScores.
It includes a sub-package for discriminative objective functions, namely conditional likelihood and supervised posterior,
and a separate sub-package for the parameter priors, that can be used for the supervised posterior.
- de.jstacs.classifiers.differentiableSequenceScoreBased.gendismix - package de.jstacs.classifiers.differentiableSequenceScoreBased.gendismix
-
Provides an implementation of a classifier that allows to train the parameters of a set of
DifferentiableStatisticalModels by
a unified generative-discriminative learning principle.
- de.jstacs.classifiers.differentiableSequenceScoreBased.logPrior - package de.jstacs.classifiers.differentiableSequenceScoreBased.logPrior
-
Provides a general definition of a parameter log-prior and a number of implementations of Laplace and Gaussian priors.
- de.jstacs.classifiers.differentiableSequenceScoreBased.msp - package de.jstacs.classifiers.differentiableSequenceScoreBased.msp
-
Provides an implementation of a classifier that allows to train the parameters of a set of
DifferentiableStatisticalModels either
by maximum supervised posterior (MSP) or by maximum conditional likelihood (MCL).
- de.jstacs.classifiers.differentiableSequenceScoreBased.sampling - package de.jstacs.classifiers.differentiableSequenceScoreBased.sampling
-
- de.jstacs.classifiers.performanceMeasures - package de.jstacs.classifiers.performanceMeasures
-
This package provides the implementations of performance measures that can be used to assess any classifier.
- de.jstacs.classifiers.trainSMBased - package de.jstacs.classifiers.trainSMBased
-
- de.jstacs.classifiers.utils - package de.jstacs.classifiers.utils
-
Provides some useful classes for working with classifiers.
- de.jstacs.clustering.distances - package de.jstacs.clustering.distances
-
- de.jstacs.clustering.hierachical - package de.jstacs.clustering.hierachical
-
- de.jstacs.data - package de.jstacs.data
-
Provides classes for the representation of data.
The base classes to represent data are Alphabet and AlphabetContainer for representing alphabets,
Sequence and its sub-classes to represent continuous and discrete sequences, and
DataSet to represent data sets comprising a set of sequences.
- de.jstacs.data.alphabets - package de.jstacs.data.alphabets
-
Provides classes for the representation of discrete and continuous alphabets, including a DNAAlphabet for the most common case of DNA-sequences.
- de.jstacs.data.bioJava - package de.jstacs.data.bioJava
-
Provides an adapter between the representation of data in BioJava and the representation used in Jstacs.
- de.jstacs.data.sequences - package de.jstacs.data.sequences
-
Provides classes for representing sequences.
The implementations of sequences currently include DiscreteSequences prepared for alphabets of different sizes, and ArbitrarySequences that may
contain continuous values as well.
As sub-package provides the facilities to annotate Sequences.
- de.jstacs.data.sequences.annotation - package de.jstacs.data.sequences.annotation
-
Provides the facilities to annotate Sequences using a number of pre-defined annotation types, or additional
implementations of the SequenceAnnotation class.
- de.jstacs.io - package de.jstacs.io
-
Provides classes for reading data from and writing to a file and storing a number of datatypes, including all primitives, arrays of primitives, and Storables to an XML-representation.
- de.jstacs.motifDiscovery - package de.jstacs.motifDiscovery
-
This package provides the framework including the interface for any de novo motif discoverer.
- de.jstacs.motifDiscovery.history - package de.jstacs.motifDiscovery.history
-
- de.jstacs.parameters - package de.jstacs.parameters
-
This package provides classes for parameters that establish a general convention for the description of parameters
as defined in the Parameter-interface.
- de.jstacs.parameters.validation - package de.jstacs.parameters.validation
-
Provides classes for the validation of Parameter values.
- de.jstacs.results - package de.jstacs.results
-
This package provides classes for results and sets of results.
- de.jstacs.results.savers - package de.jstacs.results.savers
-
- de.jstacs.sampling - package de.jstacs.sampling
-
This package contains many classes that can be used while a sampling.
- de.jstacs.sequenceScores - package de.jstacs.sequenceScores
-
Provides all
SequenceScores, which can be used to score a
Sequence, typically using some model assumptions.
- de.jstacs.sequenceScores.differentiable - package de.jstacs.sequenceScores.differentiable
-
- de.jstacs.sequenceScores.differentiable.logistic - package de.jstacs.sequenceScores.differentiable.logistic
-
- de.jstacs.sequenceScores.statisticalModels - package de.jstacs.sequenceScores.statisticalModels
-
- de.jstacs.sequenceScores.statisticalModels.differentiable - package de.jstacs.sequenceScores.statisticalModels.differentiable
-
- de.jstacs.sequenceScores.statisticalModels.differentiable.directedGraphicalModels - package de.jstacs.sequenceScores.statisticalModels.differentiable.directedGraphicalModels
-
- de.jstacs.sequenceScores.statisticalModels.differentiable.directedGraphicalModels.structureLearning.measures - package de.jstacs.sequenceScores.statisticalModels.differentiable.directedGraphicalModels.structureLearning.measures
-
- de.jstacs.sequenceScores.statisticalModels.differentiable.directedGraphicalModels.structureLearning.measures.btMeasures - package de.jstacs.sequenceScores.statisticalModels.differentiable.directedGraphicalModels.structureLearning.measures.btMeasures
-
Provides the facilities to learn the structure of a
BayesianNetworkDiffSM as
a Bayesian tree using a number of measures to define a rating of structures.
- de.jstacs.sequenceScores.statisticalModels.differentiable.directedGraphicalModels.structureLearning.measures.pmmMeasures - package de.jstacs.sequenceScores.statisticalModels.differentiable.directedGraphicalModels.structureLearning.measures.pmmMeasures
-
Provides the facilities to learn the structure of a
BayesianNetworkDiffSM as
a permuted Markov model using a number of measures to define a rating of structures.
- de.jstacs.sequenceScores.statisticalModels.differentiable.homogeneous - package de.jstacs.sequenceScores.statisticalModels.differentiable.homogeneous
-
- de.jstacs.sequenceScores.statisticalModels.differentiable.localMixture - package de.jstacs.sequenceScores.statisticalModels.differentiable.localMixture
-
- de.jstacs.sequenceScores.statisticalModels.differentiable.mixture - package de.jstacs.sequenceScores.statisticalModels.differentiable.mixture
-
- de.jstacs.sequenceScores.statisticalModels.differentiable.mixture.motif - package de.jstacs.sequenceScores.statisticalModels.differentiable.mixture.motif
-
- de.jstacs.sequenceScores.statisticalModels.trainable - package de.jstacs.sequenceScores.statisticalModels.trainable
-
- de.jstacs.sequenceScores.statisticalModels.trainable.discrete - package de.jstacs.sequenceScores.statisticalModels.trainable.discrete
-
- de.jstacs.sequenceScores.statisticalModels.trainable.discrete.homogeneous - package de.jstacs.sequenceScores.statisticalModels.trainable.discrete.homogeneous
-
- de.jstacs.sequenceScores.statisticalModels.trainable.discrete.homogeneous.parameters - package de.jstacs.sequenceScores.statisticalModels.trainable.discrete.homogeneous.parameters
-
- de.jstacs.sequenceScores.statisticalModels.trainable.discrete.inhomogeneous - package de.jstacs.sequenceScores.statisticalModels.trainable.discrete.inhomogeneous
-
This package contains various inhomogeneous models.
- de.jstacs.sequenceScores.statisticalModels.trainable.discrete.inhomogeneous.parameters - package de.jstacs.sequenceScores.statisticalModels.trainable.discrete.inhomogeneous.parameters
-
- de.jstacs.sequenceScores.statisticalModels.trainable.discrete.inhomogeneous.shared - package de.jstacs.sequenceScores.statisticalModels.trainable.discrete.inhomogeneous.shared
-
- de.jstacs.sequenceScores.statisticalModels.trainable.hmm - package de.jstacs.sequenceScores.statisticalModels.trainable.hmm
-
The package provides all interfaces and classes for a hidden Markov model (HMM).
- de.jstacs.sequenceScores.statisticalModels.trainable.hmm.models - package de.jstacs.sequenceScores.statisticalModels.trainable.hmm.models
-
The package provides different implementations of hidden Markov models based on
AbstractHMM.
- de.jstacs.sequenceScores.statisticalModels.trainable.hmm.states - package de.jstacs.sequenceScores.statisticalModels.trainable.hmm.states
-
The package provides all interfaces and classes for states used in hidden Markov models.
- de.jstacs.sequenceScores.statisticalModels.trainable.hmm.states.emissions - package de.jstacs.sequenceScores.statisticalModels.trainable.hmm.states.emissions
-
- de.jstacs.sequenceScores.statisticalModels.trainable.hmm.states.emissions.continuous - package de.jstacs.sequenceScores.statisticalModels.trainable.hmm.states.emissions.continuous
-
- de.jstacs.sequenceScores.statisticalModels.trainable.hmm.states.emissions.discrete - package de.jstacs.sequenceScores.statisticalModels.trainable.hmm.states.emissions.discrete
-
- de.jstacs.sequenceScores.statisticalModels.trainable.hmm.training - package de.jstacs.sequenceScores.statisticalModels.trainable.hmm.training
-
The package provides all classes used to determine the training algorithm of a hidden Markov model.
- de.jstacs.sequenceScores.statisticalModels.trainable.hmm.transitions - package de.jstacs.sequenceScores.statisticalModels.trainable.hmm.transitions
-
The package provides all interfaces and classes for transitions used in hidden Markov models.
- de.jstacs.sequenceScores.statisticalModels.trainable.hmm.transitions.elements - package de.jstacs.sequenceScores.statisticalModels.trainable.hmm.transitions.elements
-
- de.jstacs.sequenceScores.statisticalModels.trainable.mixture - package de.jstacs.sequenceScores.statisticalModels.trainable.mixture
-
This package is the super package for any mixture model.
- de.jstacs.sequenceScores.statisticalModels.trainable.mixture.motif - package de.jstacs.sequenceScores.statisticalModels.trainable.mixture.motif
-
- de.jstacs.sequenceScores.statisticalModels.trainable.mixture.motif.positionprior - package de.jstacs.sequenceScores.statisticalModels.trainable.mixture.motif.positionprior
-
- de.jstacs.sequenceScores.statisticalModels.trainable.phylo - package de.jstacs.sequenceScores.statisticalModels.trainable.phylo
-
- de.jstacs.sequenceScores.statisticalModels.trainable.phylo.parser - package de.jstacs.sequenceScores.statisticalModels.trainable.phylo.parser
-
- de.jstacs.tools - package de.jstacs.tools
-
- de.jstacs.tools.ui.cli - package de.jstacs.tools.ui.cli
-
- de.jstacs.tools.ui.galaxy - package de.jstacs.tools.ui.galaxy
-
- de.jstacs.utils - package de.jstacs.utils
-
This package contains a bundle of useful classes and interfaces like ...
- de.jstacs.utils.graphics - package de.jstacs.utils.graphics
-
- de.jstacs.utils.random - package de.jstacs.utils.random
-
This package contains some classes for generating random numbers.
- DeBruijnGraphSequenceGenerator - Class in de.jstacs.data
-
Class for creating De Bruin sequences using explicit De Bruijn graphs.
- DeBruijnGraphSequenceGenerator() - Constructor for class de.jstacs.data.DeBruijnGraphSequenceGenerator
-
- DeBruijnMotifComparison - Class in de.jstacs.clustering.distances
-
Helper class for comparisons of motif models based on De Bruijn sequences.
- DeBruijnMotifComparison() - Constructor for class de.jstacs.clustering.distances.DeBruijnMotifComparison
-
- DeBruijnSequenceGenerator - Class in de.jstacs.data
-
Generates De Buijn sequences using the algorithm from Frank Ruskey's Combinatorial Generation.
- DeBruijnSequenceGenerator() - Constructor for class de.jstacs.data.DeBruijnSequenceGenerator
-
- decodePath(IntList) - Method in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.AbstractHMM
-
This method decodes any path of the HMM, i.e.
- decodeStatePosterior(double[][]...) - Static method in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.AbstractHMM
-
The method returns the decoded state posterior, i.e.
- DEFAULT_INSTANCE - Static variable in class de.jstacs.data.sequences.annotation.NullSequenceAnnotationParser
-
The only instance of this class which is publicly available.
- DEFAULT_INSTANCE - Static variable in class de.jstacs.utils.random.DirichletMRG
-
This instance shall be used, since quite often two instance of this class
return the same values.
- DEFAULT_STREAM - Static variable in class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.inhomogeneous.InhomogeneousDGTrainSM
-
- DEFAULT_STREAM - Static variable in class de.jstacs.utils.SafeOutputStream
-
This stream can be used as default stream.
- defaultInstance - Static variable in class de.jstacs.classifiers.differentiableSequenceScoreBased.logPrior.DoesNothingLogPrior
-
As this prior does not penalize parameters and does not have any
parameters itself, this class does not have a constructor, but provides a
default instance in order to reduce memory consumption.
- DefaultProgressUpdater - Class in de.jstacs.utils
-
Simple class that implements
ProgressUpdater and prints the
percentage of iterations that is already done on the screen.
- DefaultProgressUpdater() - Constructor for class de.jstacs.utils.DefaultProgressUpdater
-
- defaultValue - Variable in class de.jstacs.parameters.SimpleParameter
-
The default value of the parameter
- deleteAllFilesAtTheServer() - Method in class de.jstacs.utils.REnvironment
-
Deletes all files that have been copied to the server or created on the
server.
- delta - Variable in class de.jstacs.sequenceScores.statisticalModels.differentiable.mixture.motif.DurationDiffSM
-
The difference of maximal and minimal value.
- descendants - Variable in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.transitions.BasicHigherOrderTransition.AbstractTransitionElement
-
The indices for the descendant transition elements that can be visited following the states.
- determineDiagonalElement() - Method in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.transitions.elements.ReferenceBasedTransitionElement
-
This method determines the self transition.
- determineFinalStates() - Method in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.AbstractHMM
-
This method determines the final states of the HMM.
- determineIsNormalized() - Method in class de.jstacs.sequenceScores.statisticalModels.differentiable.mixture.AbstractMixtureDiffSM
-
- DGTrainSMParameterSet<T extends DiscreteGraphicalTrainSM> - Class in de.jstacs.sequenceScores.statisticalModels.trainable.discrete
-
- DGTrainSMParameterSet(StringBuffer) - Constructor for class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.DGTrainSMParameterSet
-
The standard constructor for the interface
Storable.
- DGTrainSMParameterSet(Class<T>, boolean, boolean) - Constructor for class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.DGTrainSMParameterSet
-
An empty constructor.
- DGTrainSMParameterSet(Class<T>, AlphabetContainer, double, String) - Constructor for class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.DGTrainSMParameterSet
-
The constructor for models that can handle variable lengths.
- DGTrainSMParameterSet(Class<T>, AlphabetContainer, int, double, String) - Constructor for class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.DGTrainSMParameterSet
-
The constructor for models that can handle only sequences of fixed length
given by length.
- diagElement - Variable in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.transitions.elements.ReferenceBasedTransitionElement
-
The index of the self transition.
- diagonalWeights - Variable in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.transitions.elements.DistanceBasedScaledTransitionElement
-
Contains the single epsilons of the diagonal elements required for estimating the self-transition probability.
- diff(DataSet, DataSet...) - Static method in class de.jstacs.data.DataSet
-
This method computes the difference between the
DataSet data and
the
DataSets
samples.
- DifferentiableEmission - Interface in de.jstacs.sequenceScores.statisticalModels.trainable.hmm.states.emissions
-
This interface declares all methods needed in an emission during a numerical optimization of HMM.
- DifferentiableFunction - Class in de.jstacs.algorithms.optimization
-
This class is the framework for any (at least) one time differentiable
function
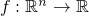
.
- DifferentiableFunction() - Constructor for class de.jstacs.algorithms.optimization.DifferentiableFunction
-
- DifferentiableHigherOrderHMM - Class in de.jstacs.sequenceScores.statisticalModels.trainable.hmm.models
-
- DifferentiableHigherOrderHMM(MaxHMMTrainingParameterSet, String[], int[], boolean[], DifferentiableEmission[], boolean, double, TransitionElement...) - Constructor for class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.models.DifferentiableHigherOrderHMM
-
This is the main constructor.
- DifferentiableHigherOrderHMM(StringBuffer) - Constructor for class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.models.DifferentiableHigherOrderHMM
-
The standard constructor for the interface
Storable.
- DifferentiableSequenceScore - Interface in de.jstacs.sequenceScores.differentiable
-
- DifferentiableState - Interface in de.jstacs.sequenceScores.statisticalModels.trainable.hmm.states
-
This interface declares a method that allows to evaluate the gradient which is essential for numerical optimization.
- DifferentiableStatisticalModel - Interface in de.jstacs.sequenceScores.statisticalModels.differentiable
-
- DifferentiableStatisticalModelFactory - Class in de.jstacs.sequenceScores.statisticalModels.differentiable
-
This class allows to easily create some frequently used models.
- DifferentiableStatisticalModelFactory() - Constructor for class de.jstacs.sequenceScores.statisticalModels.differentiable.DifferentiableStatisticalModelFactory
-
- DifferentiableStatisticalModelWrapperTrainSM - Class in de.jstacs.sequenceScores.statisticalModels.trainable
-
This model can be used to use a DifferentiableStatisticalModel as model.
- DifferentiableStatisticalModelWrapperTrainSM(DifferentiableStatisticalModel, int, byte, AbstractTerminationCondition, double, double) - Constructor for class de.jstacs.sequenceScores.statisticalModels.trainable.DifferentiableStatisticalModelWrapperTrainSM
-
The main constructor that creates an instance with the user given parameters and
CompositeLogPrior.
- DifferentiableStatisticalModelWrapperTrainSM(DifferentiableStatisticalModel, int, byte, AbstractTerminationCondition, double, double, LogPrior) - Constructor for class de.jstacs.sequenceScores.statisticalModels.trainable.DifferentiableStatisticalModelWrapperTrainSM
-
Constructor that creates an instance with the user given parameters.
- DifferentiableStatisticalModelWrapperTrainSM(StringBuffer) - Constructor for class de.jstacs.sequenceScores.statisticalModels.trainable.DifferentiableStatisticalModelWrapperTrainSM
-
The standard constructor for the interface
Storable.
- DifferentiableTransition - Interface in de.jstacs.sequenceScores.statisticalModels.trainable.hmm.transitions
-
This class declares methods that allow for optimizing the parameters numerically using the
Optimizer.
- DiffSMSamplingComponent(String) - Constructor for class de.jstacs.classifiers.differentiableSequenceScoreBased.sampling.SamplingScoreBasedClassifier.DiffSMSamplingComponent
-
- DiffSSBasedOptimizableFunction - Class in de.jstacs.classifiers.differentiableSequenceScoreBased
-
- DiffSSBasedOptimizableFunction(int, DifferentiableSequenceScore[], DataSet[], double[][], LogPrior, boolean, boolean) - Constructor for class de.jstacs.classifiers.differentiableSequenceScoreBased.DiffSSBasedOptimizableFunction
-
Creates an instance with the underlying infrastructure.
- dimension - Variable in class de.jstacs.sequenceScores.statisticalModels.trainable.mixture.AbstractMixtureTrainSM
-
The number of dimensions.
- DimensionException - Exception in de.jstacs.algorithms.optimization
-
This class is for
Exceptions depending on wrong dimensions of vectors
for a given function.
- DimensionException() - Constructor for exception de.jstacs.algorithms.optimization.DimensionException
-
Creates a new
DimensionException with standard error message
("The vector has wrong dimension for this function.").
- DimensionException(int, int) - Constructor for exception de.jstacs.algorithms.optimization.DimensionException
-
- DiMRGParams - Class in de.jstacs.utils.random
-
The super container for parameters of Dirichlet multivariate random
generators.
- DiMRGParams() - Constructor for class de.jstacs.utils.random.DiMRGParams
-
- DinucleotideProperty - Enum in de.jstacs.data
-
This enum defines physicochemical, conformational, and letter-based dinucleotide properties of nucleotide sequences.
- DinucleotideProperty.HowCreated - Enum in de.jstacs.data
-
This enum defines the origins of nucleotide properties
- DinucleotideProperty.MeanSmoothing - Class in de.jstacs.data
-
Smoothing by mean using a pre-defined window width.
- DinucleotideProperty.MedianSmoothing - Class in de.jstacs.data
-
Smoothing by median using a pre-defined window width.
- DinucleotideProperty.NoSmoothing - Class in de.jstacs.data
-
- DinucleotideProperty.Smoothing - Class in de.jstacs.data
-
Abstract class for methods that smooth a series of real values.
- DinucleotideProperty.Type - Enum in de.jstacs.data
-
This enum defines the types of dinucleotide properties.
- DirichletMRG - Class in de.jstacs.utils.random
-
This class is a multivariate random generator based on a Dirichlet
distribution.
- DirichletMRGParams - Class in de.jstacs.utils.random
-
The container for parameters of a Dirichlet random generator.
- DirichletMRGParams(double, int) - Constructor for class de.jstacs.utils.random.DirichletMRGParams
-
Constructor which creates a new hyperparameter vector for a Dirichlet
random generator.
- DirichletMRGParams(double...) - Constructor for class de.jstacs.utils.random.DirichletMRGParams
-
Constructor which creates a new hyperparameter vector for a Dirichlet
random generator.
- DirichletMRGParams(int, int, double...) - Constructor for class de.jstacs.utils.random.DirichletMRGParams
-
Constructor which creates a new hyperparameter vector for a Dirichlet
random generator.
- disconnect(AbstractList<int[]>, int[], ConstraintManager.Decomposition) - Static method in class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.ConstraintManager
-
This method tries to disconnect the constraints and create the models.
- DiscreteAlphabet - Class in de.jstacs.data.alphabets
-
Class for an alphabet that consists of arbitrary
Strings.
- DiscreteAlphabet(StringBuffer) - Constructor for class de.jstacs.data.alphabets.DiscreteAlphabet
-
The standard constructor for the interface
Storable.
- DiscreteAlphabet(DiscreteAlphabet.DiscreteAlphabetParameterSet) - Constructor for class de.jstacs.data.alphabets.DiscreteAlphabet
-
- DiscreteAlphabet(int, int) - Constructor for class de.jstacs.data.alphabets.DiscreteAlphabet
-
- DiscreteAlphabet(boolean, String...) - Constructor for class de.jstacs.data.alphabets.DiscreteAlphabet
-
- DiscreteAlphabet.DiscreteAlphabetParameterSet - Class in de.jstacs.data.alphabets
-
- DiscreteAlphabetMapping - Class in de.jstacs.data.alphabets
-
This class implements the transformation of discrete values to other discrete values
which define a
DiscreteAlphabet.
- DiscreteAlphabetMapping(int[], DiscreteAlphabet) - Constructor for class de.jstacs.data.alphabets.DiscreteAlphabetMapping
-
- DiscreteAlphabetMapping(StringBuffer) - Constructor for class de.jstacs.data.alphabets.DiscreteAlphabetMapping
-
The standard constructor for the interface
Storable.
- DiscreteAlphabetParameterSet(Class<? extends DiscreteAlphabet>) - Constructor for class de.jstacs.data.alphabets.DiscreteAlphabet.DiscreteAlphabetParameterSet
-
This constructor should only be used for parameter sets that are intended to created subclasses of
DiscreteAlphabet.
- DiscreteAlphabetParameterSet() - Constructor for class de.jstacs.data.alphabets.DiscreteAlphabet.DiscreteAlphabetParameterSet
-
- DiscreteAlphabetParameterSet(String[], boolean) - Constructor for class de.jstacs.data.alphabets.DiscreteAlphabet.DiscreteAlphabetParameterSet
-
- DiscreteAlphabetParameterSet(char[], boolean) - Constructor for class de.jstacs.data.alphabets.DiscreteAlphabet.DiscreteAlphabetParameterSet
-
- DiscreteAlphabetParameterSet(StringBuffer) - Constructor for class de.jstacs.data.alphabets.DiscreteAlphabet.DiscreteAlphabetParameterSet
-
The standard constructor for the interface
Storable
.
- DiscreteEmission - Class in de.jstacs.sequenceScores.statisticalModels.trainable.hmm.states.emissions.discrete
-
This class implements a simple discrete emission without any condition.
- DiscreteEmission(AlphabetContainer, double) - Constructor for class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.states.emissions.discrete.DiscreteEmission
-
This is a simple constructor for a
DiscreteEmission based on the equivalent sample size.
- DiscreteEmission(AlphabetContainer, double[]) - Constructor for class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.states.emissions.discrete.DiscreteEmission
-
This is a simple constructor for a
DiscreteEmission defining the individual hyper parameters.
- DiscreteEmission(StringBuffer) - Constructor for class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.states.emissions.discrete.DiscreteEmission
-
- DiscreteGraphicalTrainSM - Class in de.jstacs.sequenceScores.statisticalModels.trainable.discrete
-
This is the main class for all discrete graphical models
(DGM).
- DiscreteGraphicalTrainSM(DGTrainSMParameterSet) - Constructor for class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.DiscreteGraphicalTrainSM
-
The default constructor.
- DiscreteGraphicalTrainSM(StringBuffer) - Constructor for class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.DiscreteGraphicalTrainSM
-
The standard constructor for the interface
Storable.
- DiscreteInhomogenousDataSetEmitter - Class in de.jstacs.utils
-
Emits
DataSets for discrete inhomogeneous models by a naive implementation.
- DiscreteInhomogenousDataSetEmitter() - Constructor for class de.jstacs.utils.DiscreteInhomogenousDataSetEmitter
-
- DiscreteSequenceEnumerator - Class in de.jstacs.data
-
- DiscreteSequenceEnumerator(AlphabetContainer, int, boolean) - Constructor for class de.jstacs.data.DiscreteSequenceEnumerator
-
- discreteVal(int) - Method in class de.jstacs.data.sequences.ArbitraryFloatSequence
-
- discreteVal(int) - Method in class de.jstacs.data.sequences.ArbitrarySequence
-
- discreteVal(int) - Method in class de.jstacs.data.sequences.ByteSequence
-
- discreteVal(int) - Method in class de.jstacs.data.sequences.CyclicSequenceAdaptor
-
- discreteVal(int) - Method in class de.jstacs.data.sequences.IntSequence
-
- discreteVal(int) - Method in class de.jstacs.data.sequences.MappedDiscreteSequence
-
- discreteVal(int) - Method in class de.jstacs.data.sequences.MultiDimensionalSequence
-
- discreteVal(int) - Method in class de.jstacs.data.sequences.Sequence
-
Returns the discrete value at position
pos of the
Sequence.
- discreteVal(int) - Method in class de.jstacs.data.sequences.Sequence.RecursiveSequence
-
- discreteVal(int) - Method in class de.jstacs.data.sequences.Sequence.SubSequence
-
- discreteVal(int) - Method in class de.jstacs.data.sequences.ShortSequence
-
- discreteVal(int) - Method in class de.jstacs.data.sequences.SparseSequence
-
- discreteValAt(int) - Method in class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.inhomogeneous.SequenceIterator
-
This method returns the discrete value for a specific position.
- DistanceBasedScaledTransitionElement - Class in de.jstacs.sequenceScores.statisticalModels.trainable.hmm.transitions.elements
-
Distance-based scaled transition element for an HMM with distance-scaled transition matrices (DSHMM).
- DistanceBasedScaledTransitionElement(int[], int[], double[], double, double, String) - Constructor for class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.transitions.elements.DistanceBasedScaledTransitionElement
-
Creates an object representing the transition probabilities of a Hidden Markov TrainableStatisticalModel with scaled transition matrices (SHMM) for the given context.
- DistanceBasedScaledTransitionElement(int[], int[], double[], double, double, String, double[]) - Constructor for class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.transitions.elements.DistanceBasedScaledTransitionElement
-
Creates an object representing the transition probabilities of a Hidden Markov TrainableStatisticalModel with scaled transition matrices (SHMM) for the given context.
- DistanceBasedScaledTransitionElement(StringBuffer) - Constructor for class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.transitions.elements.DistanceBasedScaledTransitionElement
-
Extracts a distance-base scaled transition element from XML.
- DistanceMetric<T> - Class in de.jstacs.clustering.distances
-
This abstract class defined a DistanceMetric (which may be used for clustering) on a generic type T.
- DistanceMetric() - Constructor for class de.jstacs.clustering.distances.DistanceMetric
-
- divideByUnfree() - Method in class de.jstacs.sequenceScores.statisticalModels.differentiable.directedGraphicalModels.BNDiffSMParameterTree
-
Divides each of the normalized parameters on a simplex by the last
BNDiffSMParameter, which is defined not to be free.
- dList - Variable in class de.jstacs.classifiers.differentiableSequenceScoreBased.DiffSSBasedOptimizableFunction
-
These
DoubleLists are used during the parallel computation of the gradient.
- dList - Variable in class de.jstacs.sequenceScores.statisticalModels.differentiable.mixture.AbstractMixtureDiffSM
-
This array contains some
DoubleLists that are used while
computing the partial derivation.
- DNAAlphabet - Class in de.jstacs.data.alphabets
-
This class implements the discrete alphabet that is used for DNA.
- DNAAlphabet.DNAAlphabetParameterSet - Class in de.jstacs.data.alphabets
-
- DNAAlphabetContainer - Class in de.jstacs.data.alphabets
-
- DNAAlphabetContainer.DNAAlphabetContainerParameterSet - Class in de.jstacs.data.alphabets
-
- DNADataSet - Class in de.jstacs.data
-
This class exist for convenience to allow the user an easy creation of
DataSets of DNA
Sequences.
- DNADataSet(String) - Constructor for class de.jstacs.data.DNADataSet
-
Creates a new data set of DNA sequence from a FASTA file with file name fName.
- DNADataSet(String, char) - Constructor for class de.jstacs.data.DNADataSet
-
Creates a new data set of DNA sequence from a file with file name fName.
- DNADataSet(String, char, SequenceAnnotationParser) - Constructor for class de.jstacs.data.DNADataSet
-
Creates a new data set of DNA sequence from a file with file name fName using the given parser.
- document - Variable in class de.jstacs.utils.graphics.SVGAdaptor
-
The SVG document representation, may be used in sub-classes for different
Transcoders
- doesApplyFor(Sequence) - Method in class de.jstacs.sequenceScores.statisticalModels.differentiable.directedGraphicalModels.BNDiffSMParameter
-
- doesNothing() - Method in class de.jstacs.utils.SafeOutputStream
-
Indicates whether the instance is doing something or not.
- DoesNothingLogPrior - Class in de.jstacs.classifiers.differentiableSequenceScoreBased.logPrior
-
This class defines a
LogPrior that does not penalize any parameter.
- doFirstIteration(DataSet, double[]) - Method in class de.jstacs.sequenceScores.statisticalModels.trainable.mixture.AbstractMixtureTrainSM
-
This method will do the first step in the train algorithm for the current
model.
- doFirstIteration(DataSet, double[], MultivariateRandomGenerator, MRGParams[]) - Method in class de.jstacs.sequenceScores.statisticalModels.trainable.mixture.AbstractMixtureTrainSM
-
This method will do the first step in the train algorithm for the current
model.
- doFirstIteration(double[], MultivariateRandomGenerator, MRGParams[]) - Method in class de.jstacs.sequenceScores.statisticalModels.trainable.mixture.AbstractMixtureTrainSM
-
This method will do the first step in the train algorithm for the current
model on the internal data set.
- doFirstIteration(double[], MultivariateRandomGenerator, MRGParams[]) - Method in class de.jstacs.sequenceScores.statisticalModels.trainable.mixture.MixtureTrainSM
-
- doFirstIteration(DataSet, double[], double[][]) - Method in class de.jstacs.sequenceScores.statisticalModels.trainable.mixture.MixtureTrainSM
-
This method enables you to train a mixture model with a fixed start
partitioning.
- doFirstIteration(double[], MultivariateRandomGenerator, MRGParams[]) - Method in class de.jstacs.sequenceScores.statisticalModels.trainable.mixture.motif.ZOOPSTrainSM
-
- doFirstIteration(double[], MultivariateRandomGenerator, MRGParams[]) - Method in class de.jstacs.sequenceScores.statisticalModels.trainable.mixture.StrandTrainSM
-
- doHeuristicSteps(DifferentiableSequenceScore[], DataSet[], double[][], DiffSSBasedOptimizableFunction, DifferentiableFunction, byte, double, StartDistanceForecaster, SafeOutputStream, boolean, History[][], int[][], boolean) - Static method in class de.jstacs.motifDiscovery.MutableMotifDiscovererToolbox
-
- doNextIteration(int, double, double, double[], double[], double, Time) - Method in class de.jstacs.algorithms.optimization.termination.AbsoluteValueCondition
-
Deprecated.
- doNextIteration(int, double, double, double[], double[], double, Time) - Method in class de.jstacs.algorithms.optimization.termination.CombinedCondition
-
- doNextIteration(int, double, double, double[], double[], double, Time) - Method in class de.jstacs.algorithms.optimization.termination.IterationCondition
-
- doNextIteration(int, double, double, double[], double[], double, Time) - Method in class de.jstacs.algorithms.optimization.termination.MultipleIterationsCondition
-
- doNextIteration(int, double, double, double[], double[], double, Time) - Method in class de.jstacs.algorithms.optimization.termination.SmallDifferenceOfFunctionEvaluationsCondition
-
- doNextIteration(int, double, double, double[], double[], double, Time) - Method in class de.jstacs.algorithms.optimization.termination.SmallGradientConditon
-
- doNextIteration(int, double, double, double[], double[], double, Time) - Method in class de.jstacs.algorithms.optimization.termination.SmallStepCondition
-
- doNextIteration(int, double, double, double[], double[], double, Time) - Method in interface de.jstacs.algorithms.optimization.termination.TerminationCondition
-
This method allows to decide whether to do another iteration in an optimization or not.
- doNextIteration(int, double, double, double[], double[], double, Time) - Method in class de.jstacs.algorithms.optimization.termination.TimeCondition
-
- doOneSamplingStep(Function, SamplingScoreBasedClassifier.SamplingScheme, double) - Method in class de.jstacs.classifiers.differentiableSequenceScoreBased.sampling.SamplingScoreBasedClassifier
-
Performs one sampling step, i.e., one sampling of all parameter values.
- doOptimization(DataSet[], double[][]) - Method in class de.jstacs.classifiers.differentiableSequenceScoreBased.ScoreClassifier
-
This method does the optimization of the train-method
- doSingleSampling(DataSet[], double[][], int, String) - Method in class de.jstacs.classifiers.differentiableSequenceScoreBased.sampling.SamplingScoreBasedClassifier
-
Does a single sampling run for a predefined number of steps.
- DoubleArrayComparator - Class in de.jstacs.utils
-
This class implements a
Comparator of double arrays.
- DoubleArrayComparator(int) - Constructor for class de.jstacs.utils.DoubleArrayComparator
-
Creates a new instance with a specific index for comparing double arrays.
- DoubleList - Class in de.jstacs.utils
-
A simple list of primitive type double.
- DoubleList() - Constructor for class de.jstacs.utils.DoubleList
-
This is the default constructor that creates a
DoubleList with
initial length 10.
- DoubleList(int) - Constructor for class de.jstacs.utils.DoubleList
-
This is the default constructor that creates a
DoubleList with
initial length
size.
- DoubleList(StringBuffer) - Constructor for class de.jstacs.utils.DoubleList
-
This is the constructor for the interface
Storable.
- DoubleSymbolException - Exception in de.jstacs.data.alphabets
-
- DoubleSymbolException(String) - Constructor for exception de.jstacs.data.alphabets.DoubleSymbolException
-
Constructor for a
DoubleSymbolException that takes the symbol
that occurs more than once in the error message.
- DoubleTableResult(String, String, AbstractList<double[]>) - Constructor for class de.jstacs.classifiers.AbstractScoreBasedClassifier.DoubleTableResult
-
This is the default constructor that creates an instance based on the results
given in list
- DoubleTableResult(StringBuffer) - Constructor for class de.jstacs.classifiers.AbstractScoreBasedClassifier.DoubleTableResult
-
The standard constructor for the interface
Storable
.
- draw(double) - Method in class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.inhomogeneous.MEMConstraint
-
Draws the parameters from a Dirichlet.
- draw(double[], int) - Static method in class de.jstacs.sequenceScores.statisticalModels.trainable.mixture.AbstractMixtureTrainSM
-
This method draws an index of an array corresponding to the probabilities
encoded in the entries of the array.
- drawFreqs(double, InhCondProb...) - Static method in class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.ConstraintManager
-
This method draws relative frequencies.
- drawFromStatistics() - Method in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.models.SamplingHigherOrderHMM
-
This method draws all parameters for the current statistics
- drawKLDivergences(double, double[], int, int, double[][][], double) - Method in class de.jstacs.sequenceScores.statisticalModels.differentiable.directedGraphicalModels.BNDiffSMParameterTree
-
Draws KL-divergences between the distribution given by contrast and
endIdx-startIdx distributions drawn from a Dirichlet density centered around contrast, i.e.
- drawKLDivergences(double[], double[], double[][][][], double) - Method in class de.jstacs.sequenceScores.statisticalModels.differentiable.directedGraphicalModels.BNDiffSMParameterTree
-
Draws KL-divergences between the distributions given by contrast[i] each weighted by weights[i]
kls.length distributions drawn from a Dirichlet density centered around contrast, i.e.
- drawParameters(DataSet, double[]) - Method in interface de.jstacs.sampling.GibbsSamplingModel
-
This method draws the parameters of the model from the a posteriori
density.
- drawParameters(DataSet, double[]) - Method in class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.inhomogeneous.DAGTrainSM
-
This method draws the parameter of the model from the likelihood or the
posterior, respectively.
- drawParameters(DataSet, double[]) - Method in class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.inhomogeneous.FSDAGModelForGibbsSampling
-
- drawParameters(DataSet, double[], int[][]) - Method in class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.inhomogeneous.FSDAGModelForGibbsSampling
-
- drawParameters(DataSet, double[], int[][]) - Method in class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.inhomogeneous.FSDAGTrainSM
-
This method draws the parameters of the model from the a posteriori
density.
- drawParameters(double) - Method in class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.inhomogeneous.InhCondProb
-
Draws the parameters from a Dirichlet distribution using the counts and
the given ess (equivalent sample size)
as hyperparameters.
- drawParameters(double[][], boolean) - Method in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.states.emissions.discrete.AbstractConditionalDiscreteEmission
-
- drawParametersFromStatistic() - Method in interface de.jstacs.sampling.SamplingFromStatistic
-
This method draws the parameters using a sufficient statistic representing a posteriori
density.
- drawParametersFromStatistic() - Method in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.states.emissions.discrete.AbstractConditionalDiscreteEmission
-
- drawParametersFromStatistic() - Method in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.states.emissions.SilentEmission
-
- drawParametersFromStatistic() - Method in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.states.SimpleSamplingState
-
- drawParametersFromStatistic() - Method in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.transitions.BasicHigherOrderTransition.AbstractTransitionElement
-
This method draws new parameters from the sufficient statistics.
- drawParametersFromStatistic() - Method in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.transitions.BasicHigherOrderTransition
-
This method allows to draw parameters from the sufficient statistic, i.e., to draw from the posterior.
- drawPosition(int[]) - Method in class de.jstacs.sequenceScores.statisticalModels.differentiable.mixture.motif.UniformDurationDiffSM
-
This method draws from the distribution and returns the result in the given array.
- drawUnConditional(int, int, double) - Method in class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.inhomogeneous.InhCondProb
-
This method draws the parameters for a part of this constraint.
- dropBelow(IntList, S[]) - Method in class de.jstacs.clustering.hierachical.ClusterTree
-
Removes all sub-trees below the inner nodes identified by the original indexes supplied
and creates new leaf nodes instead, which obtain the supplied leaf elements.
- DualFunction(SequenceIterator, MEMConstraint[]) - Constructor for class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.inhomogeneous.MEMTools.DualFunction
-
The constructor of a dual function.
- DurationDiffSM - Class in de.jstacs.sequenceScores.statisticalModels.differentiable.mixture.motif
-
This class is the super class for all one dimensional position scoring functions that can be used as durations for semi Markov models.
- DurationDiffSM(int, int, double) - Constructor for class de.jstacs.sequenceScores.statisticalModels.differentiable.mixture.motif.DurationDiffSM
-
The default constructor.
- DurationDiffSM(StringBuffer) - Constructor for class de.jstacs.sequenceScores.statisticalModels.differentiable.mixture.motif.DurationDiffSM
-
 .
.