- e - Variable in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.states.SimpleState
-
The emission that is internally used for scoring subsequences.
- Edge - Class in de.jstacs.algorithms.graphs
-
This class is a representation of a weighted edge.
- Edge(int, int, double) - Constructor for class de.jstacs.algorithms.graphs.Edge
-
Creates a new weighted edge.
- ElementEnumerator(DataSet) - Constructor for class de.jstacs.data.DataSet.ElementEnumerator
-
- emission - Variable in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.AbstractHMM
-
The emissions used in the states.
- Emission - Interface in de.jstacs.sequenceScores.statisticalModels.trainable.hmm.states.emissions
-
This interface declares all method for an emission of a state.
- emissionIdx - Variable in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.AbstractHMM
-
The index of the used emission of each state.
- emitDataSet(int, int...) - Method in class de.jstacs.sequenceScores.statisticalModels.differentiable.AbstractDifferentiableStatisticalModel
-
- emitDataSet(int, int...) - Method in class de.jstacs.sequenceScores.statisticalModels.differentiable.directedGraphicalModels.BayesianNetworkDiffSM
-
- emitDataSet(int, int...) - Method in class de.jstacs.sequenceScores.statisticalModels.differentiable.homogeneous.HomogeneousMMDiffSM
-
This method returns a
DataSet object containing artificial
sequence(s).
- emitDataSet(int, int...) - Method in class de.jstacs.sequenceScores.statisticalModels.differentiable.IndependentProductDiffSM
-
- emitDataSet(int, int...) - Method in class de.jstacs.sequenceScores.statisticalModels.differentiable.MarkovRandomFieldDiffSM
-
- emitDataSet(int, int...) - Method in class de.jstacs.sequenceScores.statisticalModels.differentiable.mixture.MixtureDiffSM
-
- emitDataSet(int, int...) - Method in class de.jstacs.sequenceScores.statisticalModels.differentiable.UniformDiffSM
-
- emitDataSet(int, int...) - Method in interface de.jstacs.sequenceScores.statisticalModels.StatisticalModel
-
This method returns a
DataSet object containing artificial
sequence(s).
- emitDataSet(int, int...) - Method in class de.jstacs.sequenceScores.statisticalModels.trainable.AbstractTrainableStatisticalModel
-
- emitDataSet(int, int...) - Method in class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.homogeneous.HomogeneousTrainSM
-
Creates a
DataSet of a given number of
Sequences from a
trained homogeneous model.
- emitDataSet(int, int...) - Method in class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.inhomogeneous.DAGTrainSM
-
- emitDataSet(int, int...) - Method in class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.inhomogeneous.MEManager
-
- emitDataSet(int, int...) - Method in class de.jstacs.sequenceScores.statisticalModels.trainable.mixture.AbstractMixtureTrainSM
-
- emitDataSet(int, int...) - Method in class de.jstacs.sequenceScores.statisticalModels.trainable.UniformTrainSM
-
- emitDataSet(StatisticalModel, int) - Static method in class de.jstacs.utils.DiscreteInhomogenousDataSetEmitter
-
This method emits a data set with
n sequences from the discrete inhomogeneous model m
.
- emitDataSetUsingCurrentParameterSet(int, int...) - Method in class de.jstacs.sequenceScores.statisticalModels.trainable.mixture.AbstractMixtureTrainSM
-
The method returns an array of sequences using the current parameter set.
- emitDataSetUsingCurrentParameterSet(int, int...) - Method in class de.jstacs.sequenceScores.statisticalModels.trainable.mixture.MixtureTrainSM
-
- emitDataSetUsingCurrentParameterSet(int, int...) - Method in class de.jstacs.sequenceScores.statisticalModels.trainable.mixture.motif.HiddenMotifMixture
-
- emitDataSetUsingCurrentParameterSet(int, int...) - Method in class de.jstacs.sequenceScores.statisticalModels.trainable.mixture.StrandTrainSM
-
- EmptyDataSetException - Exception in de.jstacs.data
-
- EmptyDataSetException() - Constructor for exception de.jstacs.data.EmptyDataSetException
-
This constructor creates an instance with default error message
("The created DataSet is empty.").
- encode(int[][]) - Static method in class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.inhomogeneous.parameters.FSDAGTrainSMParameterSet
-
This method can be used to encode an adjacency list to a graph
description
String (e.g.
- enumerate(DifferentiableSequenceScore[], int, int, RecyclableSequenceEnumerator, double, DiffSSBasedOptimizableFunction, OutputStream) - Static method in class de.jstacs.motifDiscovery.MutableMotifDiscovererToolbox
-
This method allows to enumerate all possible seeds for a motif in the
MutableMotifDiscoverer of a specific class.
- enumerate(DifferentiableSequenceScore[], int[], int[], RecyclableSequenceEnumerator[], double, DiffSSBasedOptimizableFunction, OutputStream) - Static method in class de.jstacs.motifDiscovery.MutableMotifDiscovererToolbox
-
This method allows to enumerate all possible seeds for a number of motifs in the
MutableMotifDiscoverers of a specific classes.
- enumerateHP(Tensor) - Static method in class de.jstacs.algorithms.graphs.DAG
-
The method computes the HP(k) (see
DAG).
- EnumParameter - Class in de.jstacs.parameters
-
- EnumParameter(Class<? extends Enum>, String, boolean) - Constructor for class de.jstacs.parameters.EnumParameter
-
The main constructor.
- EnumParameter(Class<? extends Enum>, String, boolean, String) - Constructor for class de.jstacs.parameters.EnumParameter
-
This constructor creates an instance and set the default value.
- EnumParameter(StringBuffer) - Constructor for class de.jstacs.parameters.EnumParameter
-
The standard constructor for the interface
Storable.
- eps - Variable in class de.jstacs.algorithms.optimization.NumericalDifferentiableFunction
-
The constant used in the computation of the gradient.
- EPSAdaptor - Class in de.jstacs.utils.graphics
-
- EPSAdaptor() - Constructor for class de.jstacs.utils.graphics.EPSAdaptor
-
Creates a new adaptor for plotting to an EPS device.
- EqualParts - Class in de.jstacs.utils.random
-
This class is no real random generator it just returns 1/n for all values.
- EqualParts() - Constructor for class de.jstacs.utils.random.EqualParts
-
- equals(Object) - Method in class de.jstacs.data.sequences.Sequence
-
- equals(Object) - Method in class de.jstacs.parameters.AbstractSelectionParameter
-
- equals(Object) - Method in class de.jstacs.parameters.SequenceScoringParameterSet
-
- equals(Object) - Method in class de.jstacs.parameters.SimpleParameter
-
- EQUALS - Static variable in interface de.jstacs.parameters.validation.Constraint
-
The condition is equality
- equals(Object) - Method in class de.jstacs.results.SimpleResult
-
- equals(String[], String) - Static method in class de.jstacs.results.TextResult
-
Checks if the list of mime types given in p1
contains an element that is equal to one of the mime types given in mime2 (may be multiple, separated by commas).
- equals(String, String) - Static method in class de.jstacs.results.TextResult
-
Checks if the list of mime types given in mime1 (may be multiple, separated by commas)
contains an element that is equal to one of the mime types given in mime2.
- equals(Object) - Method in class de.jstacs.utils.IntList
-
- ErlangMRG - Class in de.jstacs.utils.random
-
This class is a multivariate random generator based on a Dirichlet
distribution for alpha_i \in N.
- ErlangMRG() - Constructor for class de.jstacs.utils.random.ErlangMRG
-
Constructor that creates a new multivariate random generator with
underlying Erlang distribution.
- ErlangMRGParams - Class in de.jstacs.utils.random
-
The container for parameters of an Erlang multivariate random generator.
- ErlangMRGParams(int, int) - Constructor for class de.jstacs.utils.random.ErlangMRGParams
-
Constructor which creates a new hyperparameter vector for an Erlang
random generator.
- ErlangMRGParams(int[]) - Constructor for class de.jstacs.utils.random.ErlangMRGParams
-
Constructor which creates a new hyperparameter vector for an Erlang
random generator.
- errorMessage - Variable in class de.jstacs.parameters.AbstractSelectionParameter
-
If a value was illegal for the collection parameter, this field holds the
error message.
- errorMessage - Variable in class de.jstacs.parameters.ParameterSet
-
The error message of the last error or null
- errorMessage - Variable in class de.jstacs.parameters.ParameterSetContainer
-
The message of the last error or null if no error occurred.
- ess - Variable in class de.jstacs.sequenceScores.statisticalModels.differentiable.directedGraphicalModels.BayesianNetworkDiffSM
-
The equivalent sample size.
- ess - Variable in class de.jstacs.sequenceScores.statisticalModels.differentiable.mixture.motif.DurationDiffSM
-
The equivalent sample size.
- ess - Variable in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.models.DifferentiableHigherOrderHMM
-
The equivalent sample size used for the prior
- ess - Variable in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.states.emissions.discrete.AbstractConditionalDiscreteEmission
-
The equivalent sample sizes for each condition
- ess - Variable in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.transitions.elements.ReferenceBasedTransitionElement
-
The equivalent sample size (ess) used in the prior of this instance.
- estimate(double) - Method in class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.Constraint
-
Estimates the (smoothed) relative frequencies using the ess
(equivalent sample size).
- estimate(double) - Method in class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.homogeneous.HomogeneousTrainSM.HomCondProb
-
- estimate(double) - Method in class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.inhomogeneous.InhCondProb
-
- estimate(double) - Method in class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.inhomogeneous.MEMConstraint
-
- estimateComponentProbs - Variable in class de.jstacs.sequenceScores.statisticalModels.trainable.mixture.AbstractMixtureTrainSM
-
The switch for estimating the component probabilities or not.
- estimateFromStatistic() - Method in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.states.emissions.continuous.GaussianEmission
-
- estimateFromStatistic() - Method in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.states.emissions.continuous.MultivariateGaussianEmission
-
- estimateFromStatistic() - Method in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.states.emissions.discrete.AbstractConditionalDiscreteEmission
-
- estimateFromStatistic() - Method in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.states.emissions.discrete.PhyloDiscreteEmission
-
- estimateFromStatistic() - Method in interface de.jstacs.sequenceScores.statisticalModels.trainable.hmm.states.emissions.Emission
-
This method estimates the parameters from the internal sufficient statistic.
- estimateFromStatistic() - Method in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.states.emissions.MixtureEmission
-
- estimateFromStatistic() - Method in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.states.emissions.SilentEmission
-
- estimateFromStatistic() - Method in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.states.emissions.UniformEmission
-
- estimateFromStatistic() - Method in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.transitions.BasicHigherOrderTransition.AbstractTransitionElement
-
This method estimates the parameters from the sufficient statistic.
- estimateFromStatistic() - Method in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.transitions.BasicHigherOrderTransition
-
- estimateFromStatistic() - Method in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.transitions.elements.DistanceBasedScaledTransitionElement
-
- estimateFromStatistic() - Method in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.transitions.elements.ScaledTransitionElement
-
- estimateFromStatistic() - Method in interface de.jstacs.sequenceScores.statisticalModels.trainable.hmm.transitions.TrainableTransition
-
This method estimates the parameter of the transition using the internal sufficient statistic.
- estimateFromStatistics() - Method in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.models.HigherOrderHMM
-
This method estimates the parameters of all emissions and the transition using their sufficient statistics.
- estimateParameters(DataSet, double[]) - Method in class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.inhomogeneous.DAGTrainSM
-
This method estimates the parameter of the model from the likelihood or
the posterior, respectively.
- estimateUnConditional(int, int, double, boolean) - Method in class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.Constraint
-
Estimates unconditionally.
- estimateUnConditional(int, int, double, boolean) - Method in class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.homogeneous.HomogeneousTrainSM.HomCondProb
-
- estimateUnConditional(double, double) - Method in class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.inhomogeneous.InhCondProb
-
Estimates the unconditional frequencies using the ess (equivalent
sample size).
- estimateUnConditional(int, int, double, boolean) - Method in class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.inhomogeneous.InhCondProb
-
- eval(CharSequence) - Method in class de.jstacs.utils.REnvironment
-
Evaluates the
String as R commands.
- evaluate(AbstractPerformanceMeasureParameterSet<? extends PerformanceMeasure>, boolean, DataSet...) - Method in class de.jstacs.classifiers.AbstractClassifier
-
This method evaluates the classifier and computes, for instance, the sensitivity for a given specificity, the
area under the ROC curve and so on.
- evaluate(AbstractPerformanceMeasureParameterSet<? extends PerformanceMeasure>, boolean, DataSet[], double[][]) - Method in class de.jstacs.classifiers.AbstractClassifier
-
This method evaluates the classifier and computes, for instance, the sensitivity for a given specificity, the
area under the ROC curve and so on.
- evaluateClassifier(NumericalPerformanceMeasureParameterSet, T, DataSet[], double[][], ProgressUpdater) - Method in class de.jstacs.classifiers.assessment.ClassifierAssessment
-
This method must be implemented in all subclasses.
- evaluateClassifier(NumericalPerformanceMeasureParameterSet, KFoldCrossValidationAssessParameterSet, DataSet[], double[][], ProgressUpdater) - Method in class de.jstacs.classifiers.assessment.KFoldCrossValidation
-
Evaluates a classifier.
- evaluateClassifier(NumericalPerformanceMeasureParameterSet, RepeatedHoldOutAssessParameterSet, DataSet[], double[][], ProgressUpdater) - Method in class de.jstacs.classifiers.assessment.RepeatedHoldOutExperiment
-
Evaluates the classifier.
- evaluateClassifier(NumericalPerformanceMeasureParameterSet, RepeatedSubSamplingAssessParameterSet, DataSet[], double[][], ProgressUpdater) - Method in class de.jstacs.classifiers.assessment.RepeatedSubSamplingExperiment
-
Evaluates the classifier.
- evaluateClassifier(NumericalPerformanceMeasureParameterSet, Sampled_RepeatedHoldOutAssessParameterSet, DataSet[], double[][], ProgressUpdater) - Method in class de.jstacs.classifiers.assessment.Sampled_RepeatedHoldOutExperiment
-
- evaluateFunction(double[]) - Method in interface de.jstacs.algorithms.optimization.Function
-
Evaluates the function at a certain vector (in mathematical sense)
x.
- evaluateFunction(double[]) - Method in class de.jstacs.algorithms.optimization.NegativeDifferentiableFunction
-
- evaluateFunction(double[]) - Method in class de.jstacs.algorithms.optimization.NegativeFunction
-
- evaluateFunction(double[]) - Method in class de.jstacs.algorithms.optimization.NegativeOneDimensionalFunction
-
- evaluateFunction(double) - Method in class de.jstacs.algorithms.optimization.NegativeOneDimensionalFunction
-
- evaluateFunction(double[]) - Method in class de.jstacs.algorithms.optimization.NumericalDifferentiableFunction
-
- evaluateFunction(double[]) - Method in class de.jstacs.algorithms.optimization.OneDimensionalFunction
-
- evaluateFunction(double) - Method in class de.jstacs.algorithms.optimization.OneDimensionalFunction
-
Evaluates the function at position x.
- evaluateFunction(double) - Method in class de.jstacs.algorithms.optimization.OneDimensionalSubFunction
-
- evaluateFunction(double) - Method in class de.jstacs.algorithms.optimization.QuadraticFunction
-
- evaluateFunction(double[]) - Method in class de.jstacs.classifiers.differentiableSequenceScoreBased.AbstractMultiThreadedOptimizableFunction
-
- evaluateFunction(int, int, int, int, int) - Method in class de.jstacs.classifiers.differentiableSequenceScoreBased.AbstractMultiThreadedOptimizableFunction
-
This method evaluates the function for a part of the data.
- evaluateFunction(int, int, int, int, int) - Method in class de.jstacs.classifiers.differentiableSequenceScoreBased.gendismix.LogGenDisMixFunction
-
- evaluateFunction(int, int, int, int, int) - Method in class de.jstacs.classifiers.differentiableSequenceScoreBased.gendismix.OneDataSetLogGenDisMixFunction
-
- evaluateFunction(double[]) - Method in class de.jstacs.classifiers.differentiableSequenceScoreBased.logPrior.CompositeLogPrior
-
- evaluateFunction(double[]) - Method in class de.jstacs.classifiers.differentiableSequenceScoreBased.logPrior.DoesNothingLogPrior
-
- evaluateFunction(double[]) - Method in class de.jstacs.classifiers.differentiableSequenceScoreBased.logPrior.SeparateGaussianLogPrior
-
- evaluateFunction(double[]) - Method in class de.jstacs.classifiers.differentiableSequenceScoreBased.logPrior.SeparateLaplaceLogPrior
-
- evaluateFunction(double[]) - Method in class de.jstacs.classifiers.differentiableSequenceScoreBased.logPrior.SimpleGaussianSumLogPrior
-
- evaluateFunction(double[]) - Method in class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.inhomogeneous.MEMTools.DualFunction
-
- evaluateGradientOfFunction(double[]) - Method in class de.jstacs.algorithms.optimization.DifferentiableFunction
-
Evaluates the gradient of a function at a certain vector (in mathematical
sense)
x, i.e.,
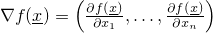
.
- evaluateGradientOfFunction(double[]) - Method in class de.jstacs.algorithms.optimization.NegativeDifferentiableFunction
-
- evaluateGradientOfFunction(double[]) - Method in class de.jstacs.algorithms.optimization.NumericalDifferentiableFunction
-
Evaluates the gradient of a function at a certain vector (in mathematical
sense) x numerically.
- evaluateGradientOfFunction(double[]) - Method in class de.jstacs.classifiers.differentiableSequenceScoreBased.AbstractMultiThreadedOptimizableFunction
-
- evaluateGradientOfFunction(int, int, int, int, int) - Method in class de.jstacs.classifiers.differentiableSequenceScoreBased.AbstractMultiThreadedOptimizableFunction
-
This method evaluates the gradient of the function for a part of the data.
- evaluateGradientOfFunction(int, int, int, int, int) - Method in class de.jstacs.classifiers.differentiableSequenceScoreBased.gendismix.LogGenDisMixFunction
-
- evaluateGradientOfFunction(int, int, int, int, int) - Method in class de.jstacs.classifiers.differentiableSequenceScoreBased.gendismix.OneDataSetLogGenDisMixFunction
-
- evaluateGradientOfFunction(double[]) - Method in class de.jstacs.classifiers.differentiableSequenceScoreBased.logPrior.LogPrior
-
- evaluateGradientOfFunction(double[]) - Method in class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.inhomogeneous.MEMTools.DualFunction
-
- EvaluationException - Exception in de.jstacs.algorithms.optimization
-
This class indicates that there was a problem to evaluate a function or the
gradient of the function.
- EvaluationException(String) - Constructor for exception de.jstacs.algorithms.optimization.EvaluationException
-
- exp - Variable in class de.jstacs.clustering.distances.SequenceScoreDistance
-
if exponential scores should be used
- ExpandableParameterSet - Class in de.jstacs.parameters
-
- ExpandableParameterSet(ParameterSet, String, String) - Constructor for class de.jstacs.parameters.ExpandableParameterSet
-
- ExpandableParameterSet(ParameterSet, String, String, int) - Constructor for class de.jstacs.parameters.ExpandableParameterSet
-
- ExpandableParameterSet(StringBuffer) - Constructor for class de.jstacs.parameters.ExpandableParameterSet
-
The standard constructor for the interface
Storable.
- ExpandableParameterSet(ParameterSet[], String, String) - Constructor for class de.jstacs.parameters.ExpandableParameterSet
-
- export(String, Result, String) - Method in class de.jstacs.tools.ui.galaxy.GalaxyAdaptor
-
Exports a specified
Result of a program execution
to a file provided by
filename and returns the
corresponding Galaxy data type.
- ExtendedZOOPSDiffSM - Class in de.jstacs.sequenceScores.statisticalModels.differentiable.mixture.motif
-
This class handles mixtures with at least one hidden motif.
- ExtendedZOOPSDiffSM(boolean, int, int, boolean, HomogeneousDiffSM, DifferentiableStatisticalModel, DurationDiffSM, boolean) - Constructor for class de.jstacs.sequenceScores.statisticalModels.differentiable.mixture.motif.ExtendedZOOPSDiffSM
-
This constructor creates an instance of
ExtendedZOOPSDiffSM that is either an OOPS or a ZOOPS model depending on the chosen
type.
- ExtendedZOOPSDiffSM(boolean, int, int, boolean, HomogeneousDiffSM, DifferentiableStatisticalModel[], DurationDiffSM[], boolean) - Constructor for class de.jstacs.sequenceScores.statisticalModels.differentiable.mixture.motif.ExtendedZOOPSDiffSM
-
- ExtendedZOOPSDiffSM(StringBuffer) - Constructor for class de.jstacs.sequenceScores.statisticalModels.differentiable.mixture.motif.ExtendedZOOPSDiffSM
-
This is the constructor for the interface
Storable.
- extendSampling(int, boolean) - Method in class de.jstacs.classifiers.differentiableSequenceScoreBased.sampling.SamplingScoreBasedClassifier.DiffSMSamplingComponent
-
- extendSampling(int, boolean) - Method in interface de.jstacs.sampling.SamplingComponent
-
This method allows to extend a sampling.
- extendSampling(int, boolean) - Method in class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.inhomogeneous.FSDAGModelForGibbsSampling
-
- extendSampling(int, boolean) - Method in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.states.emissions.discrete.AbstractConditionalDiscreteEmission
-
- extendSampling(int, boolean) - Method in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.states.emissions.SilentEmission
-
- extendSampling(int, boolean) - Method in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.states.SimpleSamplingState
-
- extendSampling(int, boolean) - Method in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.transitions.HigherOrderTransition
-
- extendSampling(int) - Method in class de.jstacs.sequenceScores.statisticalModels.trainable.mixture.AbstractMixtureTrainSM
-
This method prepares the model to extend an existing sampling.
- extract(int, String) - Static method in class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.ConstraintManager
-
Extracts the constraint of a String and returns an ArrayList of int[].
- extractAdditionalInfo(StringBuffer) - Method in class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.Constraint
-
This method parses additional information from the
StringBuffer
that is not parsed in the base class.
- extractAdditionalInfo(StringBuffer) - Method in class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.homogeneous.HomogeneousTrainSM.HomCondProb
-
- extractAdditionalInfo(StringBuffer) - Method in class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.inhomogeneous.InhCondProb
-
- extractAdditionalInfo(StringBuffer) - Method in class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.inhomogeneous.InhConstraint
-
- extractAdditionalInfo(StringBuffer) - Method in class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.inhomogeneous.MEMConstraint
-
- extractAdditionalInformation(StringBuffer) - Method in class de.jstacs.sequenceScores.statisticalModels.trainable.mixture.motif.positionprior.GaussianLikePositionPrior
-
- extractAdditionalInformation(StringBuffer) - Method in class de.jstacs.sequenceScores.statisticalModels.trainable.mixture.motif.positionprior.PositionPrior
-
This method extracts additional information from a
StringBuffer.
- extractAdditionalInformation(StringBuffer) - Method in class de.jstacs.sequenceScores.statisticalModels.trainable.mixture.motif.positionprior.UniformPositionPrior
-
- extractForTag(StringBuffer, String) - Static method in class de.jstacs.io.XMLParser
-
Extracts the contents of source between tag start and end tags.
- extractForTag(StringBuffer, String, Map<String, String>, Map<String, String>) - Static method in class de.jstacs.io.XMLParser
-
Extracts the contents of source between tag start and end tags.
- extractFurtherClassifierInfosFromXML(StringBuffer) - Method in class de.jstacs.classifiers.AbstractClassifier
-
Extracts further information of a classifier from an XML representation.
- extractFurtherClassifierInfosFromXML(StringBuffer) - Method in class de.jstacs.classifiers.AbstractScoreBasedClassifier
-
- extractFurtherClassifierInfosFromXML(StringBuffer) - Method in class de.jstacs.classifiers.differentiableSequenceScoreBased.gendismix.GenDisMixClassifier
-
- extractFurtherClassifierInfosFromXML(StringBuffer) - Method in class de.jstacs.classifiers.differentiableSequenceScoreBased.sampling.SamplingGenDisMixClassifier
-
- extractFurtherClassifierInfosFromXML(StringBuffer) - Method in class de.jstacs.classifiers.differentiableSequenceScoreBased.sampling.SamplingScoreBasedClassifier
-
- extractFurtherClassifierInfosFromXML(StringBuffer) - Method in class de.jstacs.classifiers.differentiableSequenceScoreBased.ScoreClassifier
-
- extractFurtherClassifierInfosFromXML(StringBuffer) - Method in class de.jstacs.classifiers.MappingClassifier
-
- extractFurtherClassifierInfosFromXML(StringBuffer) - Method in class de.jstacs.classifiers.trainSMBased.TrainSMBasedClassifier
-
- extractFurtherClassifierInfosFromXML(StringBuffer) - Method in class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.inhomogeneous.shared.SharedStructureClassifier
-
- extractFurtherInformation(StringBuffer) - Method in class de.jstacs.sequenceScores.differentiable.IndependentProductDiffSS
-
- extractFurtherInformation(StringBuffer) - Method in class de.jstacs.sequenceScores.differentiable.UniformDiffSS
-
- extractFurtherInformation(StringBuffer) - Method in class de.jstacs.sequenceScores.statisticalModels.differentiable.IndependentProductDiffSM
-
- extractFurtherInformation(StringBuffer) - Method in class de.jstacs.sequenceScores.statisticalModels.differentiable.mixture.AbstractMixtureDiffSM
-
- extractFurtherInformation(StringBuffer) - Method in class de.jstacs.sequenceScores.statisticalModels.differentiable.mixture.motif.ExtendedZOOPSDiffSM
-
- extractFurtherInformation(StringBuffer) - Method in class de.jstacs.sequenceScores.statisticalModels.differentiable.mixture.StrandDiffSM
-
- extractFurtherInformation(StringBuffer) - Method in class de.jstacs.sequenceScores.statisticalModels.differentiable.UniformDiffSM
-
- extractFurtherInformation(StringBuffer) - Method in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.AbstractHMM
-
This method extracts further information from the XML representation.
- extractFurtherInformation(StringBuffer) - Method in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.models.DifferentiableHigherOrderHMM
-
- extractFurtherInformation(StringBuffer) - Method in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.models.HigherOrderHMM
-
This method extracts further information from the XML representation.
- extractFurtherInformation(StringBuffer) - Method in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.models.SamplingHigherOrderHMM
-
- extractFurtherInformation(StringBuffer) - Method in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.states.emissions.discrete.AbstractConditionalDiscreteEmission
-
This method extracts further information from the XML representation.
- extractFurtherInformation(StringBuffer) - Method in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.states.emissions.discrete.PhyloDiscreteEmission
-
- extractFurtherInformation(StringBuffer) - Method in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.states.emissions.discrete.ReferenceSequenceDiscreteEmission
-
- extractFurtherInformation(StringBuffer) - Method in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.transitions.BasicHigherOrderTransition.AbstractTransitionElement
-
This method extracts further information from the XML representation.
- extractFurtherInformation(StringBuffer) - Method in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.transitions.BasicHigherOrderTransition
-
This method extracts further information from the XML representation.
- extractFurtherInformation(StringBuffer) - Method in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.transitions.elements.BasicPluginTransitionElement
-
- extractFurtherInformation(StringBuffer) - Method in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.transitions.elements.BasicTransitionElement
-
- extractFurtherInformation(StringBuffer) - Method in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.transitions.elements.DistanceBasedScaledTransitionElement
-
- extractFurtherInformation(StringBuffer) - Method in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.transitions.elements.ReferenceBasedTransitionElement
-
- extractFurtherInformation(StringBuffer) - Method in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.transitions.elements.ScaledTransitionElement
-
- extractFurtherInformation(StringBuffer) - Method in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.transitions.HigherOrderTransition
-
- extractFurtherInformation(StringBuffer) - Method in class de.jstacs.sequenceScores.statisticalModels.trainable.mixture.AbstractMixtureTrainSM
-
This method is used in the subclasses to extract further information from
the XML representation and to set these as values of the instance.
- extractFurtherInformation(StringBuffer) - Method in class de.jstacs.sequenceScores.statisticalModels.trainable.mixture.motif.HiddenMotifMixture
-
- extractFurtherInfos(StringBuffer) - Method in class de.jstacs.AnnotatedEntity
-
This method can be used in the constructor with parameter
StringBuffer to
extract the further information.
- extractFurtherInfos(StringBuffer) - Method in class de.jstacs.classifiers.AbstractScoreBasedClassifier.DoubleTableResult
-
- extractFurtherInfos(StringBuffer) - Method in class de.jstacs.parameters.AbstractSelectionParameter
-
- extractFurtherInfos(StringBuffer) - Method in class de.jstacs.parameters.EnumParameter
-
- extractFurtherInfos(StringBuffer) - Method in class de.jstacs.parameters.FileParameter
-
- extractFurtherInfos(StringBuffer) - Method in class de.jstacs.parameters.MultiSelectionParameter
-
- extractFurtherInfos(StringBuffer) - Method in class de.jstacs.parameters.Parameter
-
- extractFurtherInfos(StringBuffer) - Method in class de.jstacs.parameters.ParameterSetContainer
-
- extractFurtherInfos(StringBuffer) - Method in class de.jstacs.parameters.RangeParameter
-
- extractFurtherInfos(StringBuffer) - Method in class de.jstacs.parameters.SelectionParameter
-
- extractFurtherInfos(StringBuffer) - Method in class de.jstacs.parameters.SimpleParameter
-
- extractFurtherInfos(StringBuffer) - Method in class de.jstacs.results.DataSetResult
-
- extractFurtherInfos(StringBuffer) - Method in class de.jstacs.results.ImageResult
-
- extractFurtherInfos(StringBuffer) - Method in class de.jstacs.results.ListResult
-
- extractFurtherInfos(StringBuffer) - Method in class de.jstacs.results.PlotGeneratorResult
-
- extractFurtherInfos(StringBuffer) - Method in class de.jstacs.results.Result
-
- extractFurtherInfos(StringBuffer) - Method in class de.jstacs.results.SimpleResult
-
- extractFurtherInfos(StringBuffer) - Method in class de.jstacs.results.StorableResult
-
- extractFurtherInfos(StringBuffer) - Method in class de.jstacs.results.TextResult
-
- extractFurtherInfos(StringBuffer) - Method in class de.jstacs.tools.DataColumnParameter
-
- extractFurtherInfos(StringBuffer) - Method in class de.jstacs.tools.ToolResult
-
- extractFurtherInfos(StringBuffer) - Method in class de.jstacs.tools.ui.galaxy.GalaxyAdaptor.FileResult
-
- extractFurtherInfos(StringBuffer) - Method in class de.jstacs.tools.ui.galaxy.GalaxyAdaptor.LinkedImageResult
-
- extractObjectAndAttributesForTags(StringBuffer, String, Map<String, String>, Map<String, String>) - Static method in class de.jstacs.io.XMLParser
-
Returns the parsed value between the tags.
- extractObjectAndAttributesForTags(StringBuffer, String, Map<String, String>, Map<String, String>, Class<T>) - Static method in class de.jstacs.io.XMLParser
-
Returns the parsed value between the tags.
- extractObjectAndAttributesForTags(StringBuffer, String, Map<String, String>, Map<String, String>, Class<T>, Class<S>, S) - Static method in class de.jstacs.io.XMLParser
-
Returns the parsed value between the tags as an inner instance of the object outerInstance.
- extractObjectForTags(StringBuffer, String) - Static method in class de.jstacs.io.XMLParser
-
Returns the parsed value between the tags.
- extractObjectForTags(StringBuffer, String, Class<T>) - Static method in class de.jstacs.io.XMLParser
-
Returns the parsed value between the tags.
- extractSequenceParts(int, DataSet[], DataSet[]) - Method in class de.jstacs.sequenceScores.differentiable.IndependentProductDiffSS
-
- extractSequencesWithTags(StringBuffer, String) - Static method in class de.jstacs.io.XMLParser
-
Extracts a set of sequences from their XML representation.
- extractWeights(int, double[][]) - Method in class de.jstacs.sequenceScores.differentiable.IndependentProductDiffSS
-
 .
.