FalsePositiveRateForFixedSensitivity with empty parameter values.
FalsePositiveRateForFixedSensitivity with given sensitivity.
Storable.
Files.Parameter that represents a local file.Storable.
FileParameter.
FileParameter.
FileParameter.FileRepresentation out of the filename and the
file's contents.
FileParameter.FileRepresentation from a filename.
Storable
.
distribution.
AbstractMixtureDiffSM.componentScore with the logarithmic
scores of the components given a Sequence.
seq in a given matrix.
TransitionElement into the given array params
starting at position offset.
int[], into list.
int[], into list.
Tensor t with the weights defined in
weights.
Tensor t with the weights defined in
weights.
File using a given AlphabetContainer and a
minimal sequence length.
LinkedList of the Class objects for all classes
in listToFilter that are sub-classes of
superClass.
DinucleotidePropertys by some of their annotations.
n and does path
contraction.
pfm similar PFMs in a list of known PFMs.
T that can be instantiated, i.e.
QuadraticFunction.
x and the value
f(x), starting the search at lower.
QuadraticFunction.
sortedScores with value greater or equal to t.
T including interfaces and
abstract classes that are located in a package below
startPackage.
sortedReferenceScores and sortedMeasureScores.
Object.clone(), but does not clone the
annotation.
BNDiffSMParameterTree.
boolean indicates whether free parameterization or all
parameters are used.
command in the format defined by configBuffer of GalaxyConvertible.toGalaxy(String, String, int, StringBuffer, StringBuffer, boolean)
and sets the values of the Parameter or ParameterSet accordingly.
ParameterSet from the XML
representation as returned by ParameterSet.toXML().
ConstraintValidator from the XML representation as
returned by ConstraintValidator.toXML().
NumberValidator from the XML representation as returned
by NumberValidator.toXML().
SimpleStaticConstraint from the XML representation as
returned by SimpleStaticConstraint.toXML().
StorableValidator from the XML representation as
returned by StorableValidator.toXML().
ResultSet from its XML representation as
returned by ResultSet.toXML().
Storable
interface to create a scoring function from a StringBuffer.
StringBuffer.
AbstractHMM.AbstractHMM(StringBuffer) constructor for creating an instance from an XML representation.
GaussianEmission.GaussianEmission(StringBuffer).
AbstractConditionalDiscreteEmission.AbstractConditionalDiscreteEmission(StringBuffer).
FSDAGTrainSM) that can be used in a Gibbs sampling.Storable interface.
FSDAGModelForGibbsSampling.Storable interface.
FSDAGModelForGibbsSampling.
FSDAGTrainSM).Storable.
FSDAGTrainSM (fixed
structure directed acyclic graphical
model).Storable.
FSDAGTrainSMParameterSet set for a
FSDAGTrainSM.
FSDAGTrainSMParameterSet instance.
FSDAGTrainSMParameterSet from the
class that can be instantiated using this FSDAGTrainSMParameterSet.
FSDAGTrainSMParameterSet instance for the
specified class.
MEManager from a given
MEManagerParameterSet.
Storable.
Storable interface.
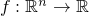 .
.DifferentiableStatisticalModels that are used to
determine the score.
DifferentiableSequenceScores using the parameters that shall be
penalized.