true if for test and train data set the sequences of
the non-reference classes have the same length as the corresponding
sequence of the reference class.
SamplingScoreBasedClassifier.burnInTest and then samples the number of
stationary steps as set in SamplingScoreBasedClassifier.params.
ClassifierAssessmentAssessParameterSet that
must be used to call the method assess( ...- Sampled_RepeatedHoldOutAssessParameterSet() -
Constructor for class de.jstacs.classifiers.assessment.Sampled_RepeatedHoldOutAssessParameterSet
- Constructs a new
Sampled_RepeatedHoldOutAssessParameterSet with
empty parameter values.
- Sampled_RepeatedHoldOutAssessParameterSet(StringBuffer) -
Constructor for class de.jstacs.classifiers.assessment.Sampled_RepeatedHoldOutAssessParameterSet
- The standard constructor for the interface
Storable.
- Sampled_RepeatedHoldOutAssessParameterSet(DataSet.PartitionMethod, int, boolean, int, int, double, boolean) -
Constructor for class de.jstacs.classifiers.assessment.Sampled_RepeatedHoldOutAssessParameterSet
- Constructs a new
Sampled_RepeatedHoldOutAssessParameterSet with
given parameter values.
- Sampled_RepeatedHoldOutExperiment - Class in de.jstacs.classifiers.assessment
- This class is a special
ClassifierAssessment that partitions the data
of a user-specified reference class (typically the smallest class) and
data sets non-overlapping for all other classes, so that one gets the same
number of sequences (and the same lengths of the sequences) in each train and
test data set. - Sampled_RepeatedHoldOutExperiment(AbstractClassifier[], TrainableStatisticalModel[][], boolean, boolean) -
Constructor for class de.jstacs.classifiers.assessment.Sampled_RepeatedHoldOutExperiment
- Creates a new
Sampled_RepeatedHoldOutExperiment from an array of
AbstractClassifiers and a two-dimensional array of TrainableStatisticalModel
s, which are combined to additional classifiers.
- Sampled_RepeatedHoldOutExperiment(AbstractClassifier...) -
Constructor for class de.jstacs.classifiers.assessment.Sampled_RepeatedHoldOutExperiment
- Creates a new
Sampled_RepeatedHoldOutExperiment from a set of
AbstractClassifiers.
- Sampled_RepeatedHoldOutExperiment(boolean, TrainableStatisticalModel[]...) -
Constructor for class de.jstacs.classifiers.assessment.Sampled_RepeatedHoldOutExperiment
- Creates a new
Sampled_RepeatedHoldOutExperiment from a set of
TrainableStatisticalModels.
- Sampled_RepeatedHoldOutExperiment(AbstractClassifier[], boolean, TrainableStatisticalModel[]...) -
Constructor for class de.jstacs.classifiers.assessment.Sampled_RepeatedHoldOutExperiment
- This constructor allows to assess a collection of given
AbstractClassifiers and those constructed using the given
TrainableStatisticalModels by a
Sampled_RepeatedHoldOutExperiment.
- sampleNSteps(DiffSSBasedOptimizableFunction, SamplingScoreBasedClassifier.DiffSMSamplingComponent, BurnInTest, int, SamplingScoreBasedClassifier.SamplingScheme) -
Method in class de.jstacs.classifiers.differentiableSequenceScoreBased.sampling.SamplingScoreBasedClassifier
- Samples a predefined number of steps appended to the current sampling
- samplePath(IntList, int, int, Sequence) -
Method in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.models.HigherOrderHMM
- This method samples a valid path for the given sequence
seq using the internal parameters.
- SamplingComponent - Interface in de.jstacs.sampling
- This interface defines methods that are used during a sampling.
- SamplingDifferentiableStatisticalModel - Interface in de.jstacs.sequenceScores.statisticalModels.differentiable
- Interface for
DifferentiableStatisticalModels that can be used for
Metropolis-Hastings sampling in a SamplingScoreBasedClassifier. - SamplingEmission - Interface in de.jstacs.sequenceScores.statisticalModels.trainable.hmm.states.emissions
-
- SamplingFromStatistic - Interface in de.jstacs.sampling
- This is the interface for sampling based on a sufficient statistic.
- SamplingGenDisMixClassifier - Class in de.jstacs.classifiers.differentiableSequenceScoreBased.sampling
- A classifier that samples its parameters from a
LogGenDisMixFunction using the
Metropolis-Hastings algorithm. - SamplingGenDisMixClassifier(SamplingGenDisMixClassifierParameterSet, BurnInTest, double[], LogPrior, double[], SamplingDifferentiableStatisticalModel...) -
Constructor for class de.jstacs.classifiers.differentiableSequenceScoreBased.sampling.SamplingGenDisMixClassifier
- Creates a new
SamplingGenDisMixClassifier using the external parameters
params, a burn-in test, a set of sampling variances for the different classes,
a prior on the parameters, weights beta for the three components of the
LogGenDisMixFunction, i.e., likelihood, conditional likelihood, and prior,
and scoring functions that model the distribution for each of the classes.
- SamplingGenDisMixClassifier(SamplingGenDisMixClassifierParameterSet, BurnInTest, double[], LogPrior, LearningPrinciple, SamplingDifferentiableStatisticalModel...) -
Constructor for class de.jstacs.classifiers.differentiableSequenceScoreBased.sampling.SamplingGenDisMixClassifier
- Creates a new
SamplingGenDisMixClassifier using the external parameters
params, a burn-in test, a set of sampling variances for the different classes,
a prior on the parameters, a learning principle,
and scoring functions that model the distribution for each of the classes.
- SamplingGenDisMixClassifier(StringBuffer) -
Constructor for class de.jstacs.classifiers.differentiableSequenceScoreBased.sampling.SamplingGenDisMixClassifier
- Creates a new
SamplingGenDisMixClassifier from its XML-representation
- SamplingGenDisMixClassifierParameterSet - Class in de.jstacs.classifiers.differentiableSequenceScoreBased.sampling
ParameterSet to instantiate a SamplingGenDisMixClassifier.- SamplingGenDisMixClassifierParameterSet(AlphabetContainer, int, int, SamplingScoreBasedClassifier.SamplingScheme, int, int, boolean, boolean, String, int) -
Constructor for class de.jstacs.classifiers.differentiableSequenceScoreBased.sampling.SamplingGenDisMixClassifierParameterSet
- Create a new
SamplingGenDisMixClassifierParameterSet.
- SamplingGenDisMixClassifierParameterSet(Class<? extends SamplingScoreBasedClassifier>, AlphabetContainer, int, int, SamplingScoreBasedClassifier.SamplingScheme, int, int, boolean, boolean, String, int) -
Constructor for class de.jstacs.classifiers.differentiableSequenceScoreBased.sampling.SamplingGenDisMixClassifierParameterSet
- Create a new
SamplingGenDisMixClassifierParameterSet.
- SamplingGenDisMixClassifierParameterSet(AlphabetContainer, int, int, int, int, String, int) -
Constructor for class de.jstacs.classifiers.differentiableSequenceScoreBased.sampling.SamplingGenDisMixClassifierParameterSet
- Create a new
SamplingGenDisMixClassifierParameterSet with a grouped sampling scheme, sampling all parameters
(and not only the free ones), and adaption of the variance.
- SamplingHigherOrderHMM - Class in de.jstacs.sequenceScores.statisticalModels.trainable.hmm.models
-
- SamplingHigherOrderHMM(SamplingHMMTrainingParameterSet, String[], int[], boolean[], SamplingEmission[], TransitionElement...) -
Constructor for class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.models.SamplingHigherOrderHMM
- This is the main constructor.
- SamplingHigherOrderHMM(StringBuffer) -
Constructor for class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.models.SamplingHigherOrderHMM
- The standard constructor for the interface
Storable.
- SamplingHigherOrderHMM.ViterbiComputation - Enum in de.jstacs.sequenceScores.statisticalModels.trainable.hmm.models
- Emumeration of all possible Viterbi-Path methods
- SamplingHMMTrainingParameterSet - Class in de.jstacs.sequenceScores.statisticalModels.trainable.hmm.training
- This class contains the parameters for training training an
AbstractHMM using a sampling strategy. - SamplingHMMTrainingParameterSet() -
Constructor for class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.training.SamplingHMMTrainingParameterSet
- This is the empty constructor that can be used to fill the parameters after creation.
- SamplingHMMTrainingParameterSet(int, int, int, AbstractBurnInTestParameterSet) -
Constructor for class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.training.SamplingHMMTrainingParameterSet
- This is the main constructor creating an already filled parameter set for training an
AbstractHMM using a sampling strategy.
- SamplingHMMTrainingParameterSet(StringBuffer) -
Constructor for class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.training.SamplingHMMTrainingParameterSet
- The standard constructor for the interface
Storable.
- samplingIndex -
Variable in class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.inhomogeneous.FSDAGModelForGibbsSampling
- The index of the current sampling.
- samplingIndex -
Variable in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.states.emissions.discrete.AbstractConditionalDiscreteEmission
- The index of the current sampling.
- samplingIndex -
Variable in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.transitions.HigherOrderTransition
- The index of the current sampling.
- samplingIndex -
Variable in class de.jstacs.sequenceScores.statisticalModels.trainable.mixture.AbstractMixtureTrainSM
- The current index of the sampling.
- SamplingPhyloHMM - Class in de.jstacs.sequenceScores.statisticalModels.trainable.hmm.models
- This class implements an (higher order) HMM that contains multi-dimensional emissions described
by a phylogenetic tree.
- SamplingPhyloHMM(SamplingHMMTrainingParameterSet, String[], int[], boolean[], PhyloDiscreteEmission[], TransitionElement...) -
Constructor for class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.models.SamplingPhyloHMM
- This is the main constructor for a hidden markov model with phylogenetic emission(s)
This model can be trained using a metropolis hastings algorithm
- SamplingPhyloHMM(StringBuffer) -
Constructor for class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.models.SamplingPhyloHMM
- The standard constructor for the interface
Storable.
- SamplingScoreBasedClassifier - Class in de.jstacs.classifiers.differentiableSequenceScoreBased.sampling
- A classifier that samples the parameters of
SamplingDifferentiableStatisticalModels by the Metropolis-Hastings algorithm. - SamplingScoreBasedClassifier(StringBuffer) -
Constructor for class de.jstacs.classifiers.differentiableSequenceScoreBased.sampling.SamplingScoreBasedClassifier
- This is the constructor for
Storable.
- SamplingScoreBasedClassifier(SamplingScoreBasedClassifierParameterSet, BurnInTest, double[], SamplingDifferentiableStatisticalModel...) -
Constructor for class de.jstacs.classifiers.differentiableSequenceScoreBased.sampling.SamplingScoreBasedClassifier
- Creates a new
SamplingScoreBasedClassifier using the parameters in params,
a specified BurnInTest (or null for no burn-in test), a set of sampling variances,
which may be different for each of the classes (in analogy to equivalent sample size for the Dirichlet distribution),
and set set of SamplingDifferentiableStatisticalModels for each of the classes.
- SamplingScoreBasedClassifier.DiffSMSamplingComponent - Class in de.jstacs.classifiers.differentiableSequenceScoreBased.sampling
- The
SamplingComponent that handles storing and loading sampled parameters values
to and from files. - SamplingScoreBasedClassifier.DiffSMSamplingComponent(String) -
Constructor for class de.jstacs.classifiers.differentiableSequenceScoreBased.sampling.SamplingScoreBasedClassifier.DiffSMSamplingComponent
- Creates a new
SamplingScoreBasedClassifier.DiffSMSamplingComponent that uses temporary files
with name prefix outfilePrefix to store sampled parameters.
- SamplingScoreBasedClassifier.SamplingScheme - Enum in de.jstacs.classifiers.differentiableSequenceScoreBased.sampling
- Sampling scheme for sampling the parameters of the scoring functions.
- SamplingScoreBasedClassifierParameterSet - Class in de.jstacs.classifiers.differentiableSequenceScoreBased.sampling
ParameterSet to instantiate a SamplingScoreBasedClassifier.- SamplingScoreBasedClassifierParameterSet(Class<? extends SamplingScoreBasedClassifier>, AlphabetContainer, int, int, SamplingScoreBasedClassifier.SamplingScheme, int, int, boolean, boolean, String) -
Constructor for class de.jstacs.classifiers.differentiableSequenceScoreBased.sampling.SamplingScoreBasedClassifierParameterSet
- Create a new
SamplingScoreBasedClassifierParameterSet.
- SamplingScoreBasedClassifierParameterSet(Class<? extends SamplingScoreBasedClassifier>, AlphabetContainer, int, int, int, int, String) -
Constructor for class de.jstacs.classifiers.differentiableSequenceScoreBased.sampling.SamplingScoreBasedClassifierParameterSet
- Create a new
SamplingScoreBasedClassifierParameterSet with a grouped sampling scheme, sampling all parameters
(and not only the free ones), and adaption of the variance.
- SamplingState - Interface in de.jstacs.sequenceScores.statisticalModels.trainable.hmm.states
-
- samplingStopped() -
Method in class de.jstacs.classifiers.differentiableSequenceScoreBased.sampling.SamplingScoreBasedClassifier.DiffSMSamplingComponent
-
- samplingStopped() -
Method in interface de.jstacs.sampling.SamplingComponent
- This method is the opposite of the method
SamplingComponent.extendSampling(int, boolean).
- samplingStopped() -
Method in class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.inhomogeneous.FSDAGModelForGibbsSampling
-
- samplingStopped() -
Method in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.states.emissions.discrete.AbstractConditionalDiscreteEmission
-
- samplingStopped() -
Method in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.states.emissions.SilentEmission
-
- samplingStopped() -
Method in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.states.SimpleSamplingState
-
- samplingStopped() -
Method in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.transitions.HigherOrderTransition
-
- samplingStopped() -
Method in class de.jstacs.sequenceScores.statisticalModels.trainable.mixture.AbstractMixtureTrainSM
- This method is the opposite of the method
AbstractMixtureTrainSM.initModelForSampling(int).
- SamplingTransition - Interface in de.jstacs.sequenceScores.statisticalModels.trainable.hmm.transitions
- This interface declares all method used during a sampling.
- satisfiesSpecificConstraint(Sequence, int) -
Method in class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.Constraint
- This method returns the index of the specific constraint that is
fulfilled by the
Sequence seq beginning at position
start.
- satisfiesSpecificConstraint(Sequence, int) -
Method in class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.homogeneous.HomogeneousTrainSM.HomCondProb
-
- satisfiesSpecificConstraint(Sequence, int) -
Method in class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.inhomogeneous.InhConstraint
-
- satisfiesSpecificConstraint(SequenceIterator) -
Method in class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.inhomogeneous.MEMConstraint
- Returns the index of the constraint that is satisfied by
sequence.
- save(File) -
Method in class de.jstacs.data.DataSet
- This method writes the
DataSet to a file f.
- save(OutputStream, char, SequenceAnnotationParser) -
Method in class de.jstacs.data.DataSet
- This method allows to write all
Sequences including their
SequenceAnnotations into a OutputStream.
- saveParameters() -
Method in class de.jstacs.classifiers.differentiableSequenceScoreBased.sampling.SamplingScoreBasedClassifier.DiffSMSamplingComponent
- Saves the parameter values of all parameter files to
a
StringBuffer representing these as XML.
- ScaledTransitionElement - Class in de.jstacs.sequenceScores.statisticalModels.trainable.hmm.transitions.elements
- Scaled transition element for an HMM with scaled transition matrices (SHMM).
- ScaledTransitionElement(int[], int[], double[], double, double[], String) -
Constructor for class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.transitions.elements.ScaledTransitionElement
- Creates an object representing the transition probabilities of a Hidden Markov TrainableStatisticalModel with scaled transition matrices (SHMM) for the given context.
- ScaledTransitionElement(StringBuffer) -
Constructor for class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.transitions.elements.ScaledTransitionElement
- The standard constructor for the interface
Storable.
- scalingFactor -
Variable in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.transitions.elements.DistanceBasedScaledTransitionElement
- The maximal scaling factor.
- scalingFactor -
Variable in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.transitions.elements.ScaledTransitionElement
- The scaling factors of the individual transition classes.
- score -
Variable in class de.jstacs.classifiers.differentiableSequenceScoreBased.DiffSSBasedOptimizableFunction
- These
DifferentiableSequenceScores are used during the parallel computation.
- score -
Variable in class de.jstacs.classifiers.differentiableSequenceScoreBased.ScoreClassifier
- The internally used scoring functions.
- score -
Variable in class de.jstacs.sequenceScores.differentiable.IndependentProductDiffSS
- The internally used
DifferentiableSequenceScores.
- score -
Variable in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.models.DifferentiableHigherOrderHMM
- The type of the score that is evaluated
- ScoreClassifier - Class in de.jstacs.classifiers.differentiableSequenceScoreBased
- This abstract class implements the main functionality of a
DifferentiableSequenceScore based classifier. - ScoreClassifier(ScoreClassifierParameterSet, double, DifferentiableSequenceScore...) -
Constructor for class de.jstacs.classifiers.differentiableSequenceScoreBased.ScoreClassifier
- Creates a new
ScoreClassifier from a given
ScoreClassifierParameterSet and DifferentiableSequenceScores .
- ScoreClassifier(StringBuffer) -
Constructor for class de.jstacs.classifiers.differentiableSequenceScoreBased.ScoreClassifier
- The standard constructor for the interface
Storable.
- ScoreClassifierParameterSet - Class in de.jstacs.classifiers.differentiableSequenceScoreBased
- A set of
Parameters for any
ScoreClassifier. - ScoreClassifierParameterSet(Class<? extends ScoreClassifier>, boolean, AlphabetContainer.AlphabetContainerType, boolean) -
Constructor for class de.jstacs.classifiers.differentiableSequenceScoreBased.ScoreClassifierParameterSet
- Creates a new
ScoreClassifierParameterSet with empty parameter
values.
- ScoreClassifierParameterSet(StringBuffer) -
Constructor for class de.jstacs.classifiers.differentiableSequenceScoreBased.ScoreClassifierParameterSet
- The standard constructor for the interface
Storable.
- ScoreClassifierParameterSet(Class<? extends ScoreClassifier>, AlphabetContainer, int, byte, double, double, double, boolean, OptimizableFunction.KindOfParameter) -
Constructor for class de.jstacs.classifiers.differentiableSequenceScoreBased.ScoreClassifierParameterSet
- The constructor for a simple, instantiated parameter set.
- ScoreClassifierParameterSet(Class<? extends ScoreClassifier>, AlphabetContainer, int, byte, AbstractTerminationCondition, double, double, boolean, OptimizableFunction.KindOfParameter) -
Constructor for class de.jstacs.classifiers.differentiableSequenceScoreBased.ScoreClassifierParameterSet
- The constructor for a simple, instantiated parameter set.
- scoringFunctions -
Variable in class de.jstacs.classifiers.differentiableSequenceScoreBased.sampling.SamplingScoreBasedClassifier
SamplingDifferentiableStatisticalModels
- sd(int, int) -
Method in class de.jstacs.utils.DoubleList
- This method computes the standard deviation of a part of the list.
- sd(int, int, double[]) -
Static method in class de.jstacs.utils.ToolBox
- This method returns the standard deviation of the elements of an
array
between start and end.
- SelectionParameter - Class in de.jstacs.parameters
- Class for a collection parameter, i.e.
- SelectionParameter(DataType, String[], Object[], String, String, boolean) -
Constructor for class de.jstacs.parameters.SelectionParameter
- Constructor for a
SelectionParameter.
- SelectionParameter(DataType, String[], Object[], String[], String, String, boolean) -
Constructor for class de.jstacs.parameters.SelectionParameter
- Constructor for a
SelectionParameter.
- SelectionParameter(String, String, boolean, ParameterSet...) -
Constructor for class de.jstacs.parameters.SelectionParameter
- Constructor for a
SelectionParameter from an array of
ParameterSets.
- SelectionParameter(String, String, boolean, Class<? extends ParameterSet>...) -
Constructor for class de.jstacs.parameters.SelectionParameter
- Constructor for a
SelectionParameter from an array of
Classes of ParameterSets.
- SelectionParameter(StringBuffer) -
Constructor for class de.jstacs.parameters.SelectionParameter
- The standard constructor for the interface
Storable.
- SensitivityForFixedSpecificity - Class in de.jstacs.classifiers.performanceMeasures
- This class implements the sensitivity for a fixed specificity.
- SensitivityForFixedSpecificity() -
Constructor for class de.jstacs.classifiers.performanceMeasures.SensitivityForFixedSpecificity
- Constructs a new instance of the performance measure
SensitivityForFixedSpecificity with empty parameter values.
- SensitivityForFixedSpecificity(double) -
Constructor for class de.jstacs.classifiers.performanceMeasures.SensitivityForFixedSpecificity
- Constructs a new instance of the performance measure
SensitivityForFixedSpecificity with given specificity.
- SensitivityForFixedSpecificity(StringBuffer) -
Constructor for class de.jstacs.classifiers.performanceMeasures.SensitivityForFixedSpecificity
- The standard constructor for the interface
Storable.
- SeparateGaussianLogPrior - Class in de.jstacs.classifiers.differentiableSequenceScoreBased.logPrior
- Class for a
LogPrior that defines a Gaussian prior on the parameters
of a set of DifferentiableStatisticalModels
and a set of class parameters. - SeparateGaussianLogPrior(double[], double[], double[]) -
Constructor for class de.jstacs.classifiers.differentiableSequenceScoreBased.logPrior.SeparateGaussianLogPrior
- Creates a new
SeparateGaussianLogPrior from a set of base
variances vars, a set of class variances
classVars and a set of class means classMus.
- SeparateGaussianLogPrior(StringBuffer) -
Constructor for class de.jstacs.classifiers.differentiableSequenceScoreBased.logPrior.SeparateGaussianLogPrior
- The standard constructor for the interface
Storable.
- SeparateLaplaceLogPrior - Class in de.jstacs.classifiers.differentiableSequenceScoreBased.logPrior
- Class for a
LogPrior that defines a Laplace prior on the parameters
of a set of DifferentiableStatisticalModels
and a set of class parameters. - SeparateLaplaceLogPrior(double[], double[], double[]) -
Constructor for class de.jstacs.classifiers.differentiableSequenceScoreBased.logPrior.SeparateLaplaceLogPrior
- Creates a new
SeparateLaplaceLogPrior from a set of base
variances vars, a set of class variances
classVars and a set of class means classMus.
- SeparateLaplaceLogPrior(StringBuffer) -
Constructor for class de.jstacs.classifiers.differentiableSequenceScoreBased.logPrior.SeparateLaplaceLogPrior
- The standard constructor for the interface
Storable.
- SeparateLogPrior - Class in de.jstacs.classifiers.differentiableSequenceScoreBased.logPrior
- Abstract class for priors that penalize each parameter value independently
and have some variances (and possible means) as hyperparameters.
- SeparateLogPrior(double[], double[], double[]) -
Constructor for class de.jstacs.classifiers.differentiableSequenceScoreBased.logPrior.SeparateLogPrior
- Creates a new
SeparateLogPrior using the class-specific base
variances vars, the variances classVars and the
means classMus for the class parameters.
- SeparateLogPrior(StringBuffer) -
Constructor for class de.jstacs.classifiers.differentiableSequenceScoreBased.logPrior.SeparateLogPrior
- The standard constructor for the interface
Storable.
- SeqLogoPlotter - Class in de.jstacs.utils
- Class with static methods for plotting sequence logos of DNA motifs, i.e., position weight matrices defined over a
DNAAlphabet. - SeqLogoPlotter() -
Constructor for class de.jstacs.utils.SeqLogoPlotter
-
- Sequence<T> - Class in de.jstacs.data.sequences
- This is the main class for all sequences.
- Sequence(AlphabetContainer, SequenceAnnotation[]) -
Constructor for class de.jstacs.data.sequences.Sequence
- Creates a new
Sequence with the given AlphabetContainer
and the given annotation, but without the content.
- Sequence.CompositeSequence<T> - Class in de.jstacs.data.sequences
- The class handles composite
Sequences. - Sequence.CompositeSequence(Sequence, int[], int[]) -
Constructor for class de.jstacs.data.sequences.Sequence.CompositeSequence
- This is a very efficient way to create a
Sequence.CompositeSequence
for Sequences with a simple AlphabetContainer.
- Sequence.CompositeSequence(AlphabetContainer, Sequence<T>, int[], int[]) -
Constructor for class de.jstacs.data.sequences.Sequence.CompositeSequence
- This constructor should be used if one wants to create a
DataSet of Sequence.CompositeSequences.
- Sequence.RecursiveSequence<T> - Class in de.jstacs.data.sequences
- This is the main class for subsequences, composite sequences, ...
- Sequence.RecursiveSequence(AlphabetContainer, SequenceAnnotation[], Sequence<T>) -
Constructor for class de.jstacs.data.sequences.Sequence.RecursiveSequence
- Creates a new
Sequence.RecursiveSequence on the Sequence
seq with the AlphabetContainer alphabet
and the annotation annotation.
- Sequence.RecursiveSequence(AlphabetContainer, Sequence<T>) -
Constructor for class de.jstacs.data.sequences.Sequence.RecursiveSequence
- Creates a new
Sequence.RecursiveSequence on the Sequence
seq with the AlphabetContainer alphabet
using the annotation of the given Sequence.
- Sequence.SubSequence<T> - Class in de.jstacs.data.sequences
- This class handles subsequences.
- Sequence.SubSequence(AlphabetContainer, Sequence, int, int) -
Constructor for class de.jstacs.data.sequences.Sequence.SubSequence
- This constructor should be used if one wants to create a
DataSet of Sequence.SubSequences of defined length.
- Sequence.SubSequence(Sequence, int, int) -
Constructor for class de.jstacs.data.sequences.Sequence.SubSequence
- This is a very efficient way to create a
Sequence.SubSequence of
defined length for Sequences with a simple
AlphabetContainer.
- SequenceAnnotation - Class in de.jstacs.data.sequences.annotation
- Class for a general annotation of a
Sequence. - SequenceAnnotation(String, String, Result) -
Constructor for class de.jstacs.data.sequences.annotation.SequenceAnnotation
- Creates a new
SequenceAnnotation of type type with
identifier identifier and additional annotation (that does
not fit the SequenceAnnotation definitions) given as a
Result result.
- SequenceAnnotation(String, String, Result[]...) -
Constructor for class de.jstacs.data.sequences.annotation.SequenceAnnotation
- Creates a new
SequenceAnnotation of type type with
identifier identifier and additional annotation (that does
not fit the SequenceAnnotation definitions) given as an array of
Results results.
- SequenceAnnotation(String, String, SequenceAnnotation[], Result...) -
Constructor for class de.jstacs.data.sequences.annotation.SequenceAnnotation
- Creates a new
SequenceAnnotation of type type with
identifier identifier and additional annotation (that does
not fit the SequenceAnnotation definitions) given as an array of
Results additionalAnnotation.
- SequenceAnnotation(String, String, Collection<? extends Result>) -
Constructor for class de.jstacs.data.sequences.annotation.SequenceAnnotation
- Creates a new
SequenceAnnotation of type type with
identifier identifier and additional annotation (that does
not fit the SequenceAnnotation definitions) given as a
Collection of Results results.
- SequenceAnnotation(StringBuffer) -
Constructor for class de.jstacs.data.sequences.annotation.SequenceAnnotation
- The standard constructor for the interface
Storable.
- SequenceAnnotationParser - Interface in de.jstacs.data.sequences.annotation
- This interface declares the methods which are used by
AbstractStringExtractor to annotate a String
which will be parsed to a Sequence. - SequenceEnumeration - Class in de.jstacs.data
- This class implements a
RecyclableSequenceEnumerator on user-specified Sequences. - SequenceEnumeration(Sequence...) -
Constructor for class de.jstacs.data.SequenceEnumeration
- This constructor creates an instance based on the user-specified
Sequences sequences.
- SequenceEnumeration(Collection<Sequence>) -
Constructor for class de.jstacs.data.SequenceEnumeration
- This constructor creates an instance based on the user-specified
Collection of Sequences sequences.
- SequenceIterator - Class in de.jstacs.sequenceScores.statisticalModels.trainable.discrete.inhomogeneous
- This class is used to iterate over a discrete sequence.
- SequenceIterator(int) -
Constructor for class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.inhomogeneous.SequenceIterator
- Creates a new
SequenceIterator with maximal length.
- sequenceIteratorToDataSet(SequenceIterator, FeatureFilter) -
Static method in class de.jstacs.data.bioJava.BioJavaAdapter
- This method creates a new
DataSet from a SequenceIterator.
- SequenceScore - Interface in de.jstacs.sequenceScores
- This interface defines a scoring function that assigns a score to each input sequence.
- SequenceScoringParameterSet<T extends InstantiableFromParameterSet> - Class in de.jstacs.parameters
- Abstract class for a
ParameterSet containing all parameters necessary
to construct an Object that implements
InstantiableFromParameterSet. - SequenceScoringParameterSet(Class<T>, AlphabetContainer.AlphabetContainerType, boolean) -
Constructor for class de.jstacs.parameters.SequenceScoringParameterSet
- Constructs an
InstanceParameterSet having empty parameter values.
- SequenceScoringParameterSet(Class<T>, AlphabetContainer.AlphabetContainerType, boolean, boolean) -
Constructor for class de.jstacs.parameters.SequenceScoringParameterSet
- Constructs a
SequenceScoringParameterSet having empty parameter
values.
- SequenceScoringParameterSet(StringBuffer) -
Constructor for class de.jstacs.parameters.SequenceScoringParameterSet
- The standard constructor for the interface
Storable.
- SequenceScoringParameterSet(Class<T>, AlphabetContainer, int, boolean) -
Constructor for class de.jstacs.parameters.SequenceScoringParameterSet
- Constructs a
SequenceScoringParameterSet from an
AlphabetContainer and the length of a sequence.
- SequenceScoringParameterSet(Class<T>, AlphabetContainer) -
Constructor for class de.jstacs.parameters.SequenceScoringParameterSet
- Constructs a
SequenceScoringParameterSet for an object that can
handle sequences of variable length and with the
AlphabetContainer alphabet.
- seqWeights -
Variable in class de.jstacs.sequenceScores.statisticalModels.trainable.mixture.AbstractMixtureTrainSM
- The weights of the (sub-)sequence used to train the components (internal models).
- set() -
Method in class de.jstacs.algorithms.optimization.termination.AbsoluteValueCondition
- Deprecated.
- set() -
Method in class de.jstacs.algorithms.optimization.termination.AbstractTerminationCondition
- This method sets internal member variables from
AbstractTerminationCondition.parameter.
- set() -
Method in class de.jstacs.algorithms.optimization.termination.CombinedCondition
-
- set() -
Method in class de.jstacs.algorithms.optimization.termination.IterationCondition
-
- set() -
Method in class de.jstacs.algorithms.optimization.termination.MultipleIterationsCondition
-
- set() -
Method in class de.jstacs.algorithms.optimization.termination.SmallDifferenceOfFunctionEvaluationsCondition
-
- set() -
Method in class de.jstacs.algorithms.optimization.termination.SmallGradientConditon
-
- set() -
Method in class de.jstacs.algorithms.optimization.termination.SmallStepCondition
-
- set() -
Method in class de.jstacs.algorithms.optimization.termination.TimeCondition
-
- set(int, T) -
Method in class de.jstacs.AnnotatedEntityList
- Replaces the
AnnotatedEntity at index idx with
the AnnotatedEntity entity/code>
- set(boolean, DifferentiableSequenceScore...) -
Method in class de.jstacs.classifiers.differentiableSequenceScoreBased.logPrior.CompositeLogPrior
-
- set(boolean, DifferentiableSequenceScore...) -
Method in class de.jstacs.classifiers.differentiableSequenceScoreBased.logPrior.LogPrior
- Resets all pre-computed values to their initial values using the
DifferentiableSequenceScores funs.
- set(boolean, DifferentiableSequenceScore...) -
Method in class de.jstacs.classifiers.differentiableSequenceScoreBased.logPrior.SeparateLogPrior
-
- set(int, Parameter) -
Method in class de.jstacs.parameters.ParameterSet.ParameterList
-
- set(DGTrainSMParameterSet, boolean) -
Method in class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.DiscreteGraphicalTrainSM
- Sets the parameters as internal parameters and does some essential
computations.
- set(DGTrainSMParameterSet, boolean) -
Method in class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.homogeneous.HomogeneousMM
-
- set(DGTrainSMParameterSet, boolean) -
Method in class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.homogeneous.HomogeneousTrainSM
-
- set(DGTrainSMParameterSet, boolean) -
Method in class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.inhomogeneous.BayesianNetworkTrainSM
-
- set(DGTrainSMParameterSet, boolean) -
Method in class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.inhomogeneous.FSDAGTrainSM
-
- set(DGTrainSMParameterSet, boolean) -
Method in class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.inhomogeneous.FSMEManager
-
- set(DGTrainSMParameterSet, boolean) -
Method in class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.inhomogeneous.InhomogeneousDGTrainSM
-
- setAlpha(double) -
Method in class de.jstacs.sequenceScores.statisticalModels.trainable.mixture.AbstractMixtureTrainSM
- Sets the parameter of the Dirichlet distribution which is used when you
invoke
train to init the gammas.
- setBounds(int[]) -
Method in class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.inhomogeneous.SequenceIterator
- This method sets the bounds for each position.
- setClassWeights(boolean, double...) -
Method in class de.jstacs.classifiers.AbstractScoreBasedClassifier
- Sets new class weights.
- setClassWeights(boolean, double[], int) -
Method in class de.jstacs.classifiers.AbstractScoreBasedClassifier
- Sets new class weights.
- setCurrentLength(int) -
Method in class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.inhomogeneous.CombinationIterator
- This method sets the current used number of selected elements.
- setCurrentSamplingIndex(int) -
Method in class de.jstacs.sampling.AbstractBurnInTest
-
- setCurrentSamplingIndex(int) -
Method in interface de.jstacs.sampling.BurnInTest
- This method sets the value of the current sampling.
- setCurrentSamplingIndex(int) -
Method in class de.jstacs.sampling.SimpleBurnInTest
- Deprecated.
- setDataAndWeights(DataSet[], double[][]) -
Method in class de.jstacs.classifiers.differentiableSequenceScoreBased.AbstractMultiThreadedOptimizableFunction
-
- setDataAndWeights(DataSet[], double[][]) -
Method in class de.jstacs.classifiers.differentiableSequenceScoreBased.AbstractOptimizableFunction
-
- setDataAndWeights(DataSet[], double[][]) -
Method in class de.jstacs.classifiers.differentiableSequenceScoreBased.gendismix.OneDataSetLogGenDisMixFunction
-
- setDataAndWeights(DataSet[], double[][]) -
Method in class de.jstacs.classifiers.differentiableSequenceScoreBased.OptimizableFunction
- This method sets the data set and the sequence weights to be used.
- setDefault(Object) -
Method in class de.jstacs.parameters.AbstractSelectionParameter
- Sets the default value of this
AbstractSelectionParameter to
defaultValue.
- setDefault(Object) -
Method in class de.jstacs.parameters.EnumParameter
-
- setDefault(Object) -
Method in class de.jstacs.parameters.FileParameter
-
- setDefault(Object) -
Method in class de.jstacs.parameters.MultiSelectionParameter
-
- setDefault(Object) -
Method in class de.jstacs.parameters.Parameter
- Sets the default value of the
Parameter to
defaultValue.
- setDefault(Object) -
Method in class de.jstacs.parameters.ParameterSetContainer
-
- setDefault(Object) -
Method in class de.jstacs.parameters.RangeParameter
-
- setDefault(Object) -
Method in class de.jstacs.parameters.SelectionParameter
-
- setDefault(Object) -
Method in class de.jstacs.parameters.SimpleParameter
-
- setDefaultSelected(int[]) -
Method in class de.jstacs.parameters.MultiSelectionParameter
- Sets the default selection of this
MultiSelectionParameter to
defaultSelection.
- setDeleteOnExit(boolean) -
Method in class de.jstacs.classifiers.differentiableSequenceScoreBased.sampling.SamplingScoreBasedClassifier
- If set to
true (which is the default), the temporary files for storing sampled parameter
values are deleted on exit of the program.
- setEss(double) -
Method in class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.DGTrainSMParameterSet
- This method can be used to set the ess (equivalent sample
size) of this parameter set.
- setESS(double) -
Method in class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.inhomogeneous.StructureLearner
- This method sets the ess (equivalent sample size) of
the
StructureLearner.
- setExpLambda(int, double) -
Method in class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.inhomogeneous.MEMConstraint
- Sets the exponential value of
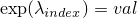 .
.
(Additionally it sets the value of 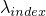 to the logarithmic value of
to the logarithmic value of val:
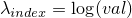 .)
.)
- setExtension(String) -
Method in class de.jstacs.parameters.FileParameter.FileRepresentation
- Sets the extension of this
FileParameter.
- setExtension(String) -
Method in class de.jstacs.utils.galaxy.GalaxyAdaptor.FileResult
- Sets the filename extension
- setFilename(String) -
Method in class de.jstacs.utils.galaxy.GalaxyAdaptor.FileResult
- Sets the file
- setForwardProb(double) -
Method in class de.jstacs.sequenceScores.statisticalModels.differentiable.mixture.StrandDiffSM
- This method can be used to set the forward strand probability.
- setFrameParameterOptimization(boolean) -
Method in class de.jstacs.sequenceScores.statisticalModels.differentiable.CyclicMarkovModelDiffSM
- This method enables the user to choose whether the frame parameters should be optimized or not.
- setFreqs(String[], int) -
Method in class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.inhomogeneous.InhCondProb
- This method is used to restore the values of a Gibbs Sampling run.
- setFurtherInformation(StringBuffer) -
Method in class de.jstacs.sampling.AbstractBurnInTest
- This method sets further information for the
AbstractBurnInTest.
- setFurtherInformation(StringBuffer) -
Method in class de.jstacs.sampling.VarianceRatioBurnInTest
-
- setFurtherModelInfos(StringBuffer) -
Method in class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.DiscreteGraphicalTrainSM
- This method replaces the internal model information with those from a
StringBuffer.
- setFurtherModelInfos(StringBuffer) -
Method in class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.homogeneous.HomogeneousMM
-
- setFurtherModelInfos(StringBuffer) -
Method in class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.inhomogeneous.DAGTrainSM
-
- setFurtherModelInfos(StringBuffer) -
Method in class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.inhomogeneous.FSDAGModelForGibbsSampling
-
- setFurtherModelInfos(StringBuffer) -
Method in class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.inhomogeneous.MEManager
-
- setHelp(String) -
Method in class de.jstacs.utils.galaxy.GalaxyAdaptor
- Sets the help, i.e., a more detailed description of the program
to
help.
- setHelp(File) -
Method in class de.jstacs.utils.galaxy.GalaxyAdaptor
- Sets the help, i.e., a more detailed description of the program
to the contents of
helpfile.
- setHiddenParameters(double[], int) -
Method in class de.jstacs.sequenceScores.statisticalModels.differentiable.mixture.AbstractMixtureDiffSM
- This method sets the hidden parameters of the model.
- setId(int) -
Method in class de.jstacs.sequenceScores.statisticalModels.trainable.phylo.PhyloNode
- This method set the ID of the current PhyloNode
The ID should be unique in the PhyloTree
- setIndexOfDescendantTransitionElement(int, int) -
Method in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.transitions.BasicHigherOrderTransition.AbstractTransitionElement
- This method sets the index of the descendant transition element for the child with index
index.
- setInitParameters(double[]) -
Method in class de.jstacs.classifiers.differentiableSequenceScoreBased.sampling.SamplingScoreBasedClassifier
- Sets the initial parameters of the sampling to
parameters.
- setLambda(int, double) -
Method in class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.inhomogeneous.MEMConstraint
- Sets the value of
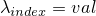 .
.
(Additionally it sets the exponential value of 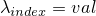 to the exponential value of
to the exponential value of val:
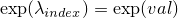 .
.
- setLastDistance(double) -
Method in class de.jstacs.algorithms.optimization.ConstantStartDistance
-
- setLastDistance(double) -
Method in class de.jstacs.algorithms.optimization.LimitedMedianStartDistance
-
- setLastDistance(double) -
Method in interface de.jstacs.algorithms.optimization.StartDistanceForecaster
- Sets the last used distance.
- setLinear(boolean) -
Method in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.states.emissions.discrete.AbstractConditionalDiscreteEmission
- If set to true, the probabilities are mapped to colors by directly, otherwise
a logistic mapping is used to emphasize deviations from the uniform distribution.
- setMax(int) -
Method in class de.jstacs.utils.DefaultProgressUpdater
-
- setMax(int) -
Method in class de.jstacs.utils.GUIProgressUpdater
-
- setMax(int) -
Method in class de.jstacs.utils.NullProgressUpdater
-
- setMax(int) -
Method in interface de.jstacs.utils.ProgressUpdater
- Sets the maximal value that will be set by
ProgressUpdater.setValue(int), so a
value of max indicates the end of the supervised method call.
- setMeasure(T) -
Method in class de.jstacs.classifiers.performanceMeasures.AbstractPerformanceMeasureParameterSet
- Sets the given measure as content of the internally last
ParameterSetContainer.
- setModelType(String) -
Method in class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.inhomogeneous.parameters.BayesianNetworkTrainSMParameterSet
- This method allows a simple change of the model type.
- setMotifLength(int) -
Method in class de.jstacs.sequenceScores.statisticalModels.trainable.mixture.motif.positionprior.GaussianLikePositionPrior
-
- setMotifLength(int) -
Method in class de.jstacs.sequenceScores.statisticalModels.trainable.mixture.motif.positionprior.PositionPrior
- Sets the length of the current motif.
- setName(String) -
Method in class de.jstacs.sequenceScores.statisticalModels.trainable.phylo.PhyloNode
- This method set a name for the current instance
- setNumberOfStarts(int) -
Method in class de.jstacs.classifiers.differentiableSequenceScoreBased.sampling.SamplingScoreBasedClassifierParameterSet
- Sets the number of starts to
i
- setNumberOfThreads(int) -
Method in class de.jstacs.classifiers.differentiableSequenceScoreBased.gendismix.GenDisMixClassifier
- This method allows to set the number of threads used while optimization.
- setNumberOfThreads(int) -
Method in class de.jstacs.classifiers.differentiableSequenceScoreBased.gendismix.GenDisMixClassifierParameterSet
- This method set the number of threads used during optimization.
- setOffset() -
Method in class de.jstacs.utils.NullProgressUpdater
- After
NullProgressUpdater.setOffset() is called the current value
will be added to every value set by
NullProgressUpdater.setValue(int).
- setOutputStream(OutputStream) -
Method in class de.jstacs.classifiers.differentiableSequenceScoreBased.ScoreClassifier
- Sets the
OutputStream that is used e.g.
- setOutputStream(OutputStream) -
Method in class de.jstacs.sequenceScores.statisticalModels.trainable.DifferentiableStatisticalModelWrapperTrainSM
- Sets the OutputStream that is used e.g.
- setOutputStream(OutputStream) -
Method in class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.inhomogeneous.InhomogeneousDGTrainSM
- Sets the
OutputStream for the model.
- setOutputStream(OutputStream) -
Method in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.AbstractHMM
- Sets the
OutputStream that is used e.g.
- setOutputStream(OutputStream) -
Method in class de.jstacs.sequenceScores.statisticalModels.trainable.mixture.AbstractMixtureTrainSM
- Sets the
OutputStream that is used e.g.
- setParameter(double[], int) -
Method in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.states.emissions.continuous.GaussianEmission
-
- setParameter(double[], int) -
Method in interface de.jstacs.sequenceScores.statisticalModels.trainable.hmm.states.emissions.DifferentiableEmission
- This method sets the internal parameters using the given global parameter array, the global offset of the HMM and the internal offset.
- setParameter(double[], int) -
Method in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.states.emissions.discrete.AbstractConditionalDiscreteEmission
-
- setParameter(double[], int) -
Method in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.states.emissions.SilentEmission
-
- setParameter(double[], int) -
Method in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.states.emissions.UniformEmission
-
- setParameterFor(int, int[][], BNDiffSMParameter) -
Method in class de.jstacs.sequenceScores.statisticalModels.differentiable.directedGraphicalModels.BNDiffSMParameterTree
- Sets the instance of the
BNDiffSMParameter for symbol symbol and
context context to BNDiffSMParameter par.
- setParameterOffset(int) -
Method in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.states.emissions.continuous.GaussianEmission
-
- setParameterOffset(int) -
Method in interface de.jstacs.sequenceScores.statisticalModels.trainable.hmm.states.emissions.DifferentiableEmission
- This method sets the internal parameter offset and returns the new parameter offset for further use.
- setParameterOffset(int) -
Method in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.states.emissions.discrete.AbstractConditionalDiscreteEmission
-
- setParameterOffset(int) -
Method in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.states.emissions.SilentEmission
-
- setParameterOffset(int) -
Method in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.states.emissions.UniformEmission
-
- setParameterOffset(int) -
Method in interface de.jstacs.sequenceScores.statisticalModels.trainable.hmm.transitions.DifferentiableTransition
- This method sets the internal offset of the parameter index.
- setParameterOffset(int) -
Method in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.transitions.elements.TransitionElement
- This method sets the internal
TransitionElement.offset used for several methods (cf.
- setParameterOffset(int) -
Method in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.transitions.HigherOrderTransition
-
- setParameterOffset() -
Method in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.transitions.HigherOrderTransition
- This method allows to set the parameter offset in each internally used
TransitionElement.
- setParameterOptimization(boolean) -
Method in class de.jstacs.sequenceScores.statisticalModels.differentiable.CyclicMarkovModelDiffSM
- This method enables the user to choose whether the parameters should be optimized or not.
- setParameterOptimization(boolean) -
Method in class de.jstacs.sequenceScores.statisticalModels.differentiable.homogeneous.HomogeneousMMDiffSM
- This method allows the user to specify whether the parameters should be
optimized or not.
- setParameters(double[], int) -
Method in interface de.jstacs.sequenceScores.differentiable.DifferentiableSequenceScore
- This method sets the internal parameters to the values of
params between start and
start + DifferentiableSequenceScore.getNumberOfParameters() - 1
- setParameters(double[], int) -
Method in class de.jstacs.sequenceScores.differentiable.IndependentProductDiffSS
-
- setParameters(double[], int) -
Method in class de.jstacs.sequenceScores.differentiable.logistic.LogisticDiffSS
-
- setParameters(double[], int) -
Method in class de.jstacs.sequenceScores.differentiable.MultiDimensionalSequenceWrapperDiffSS
-
- setParameters(double[], int) -
Method in class de.jstacs.sequenceScores.differentiable.UniformDiffSS
-
- setParameters(double[], int) -
Method in class de.jstacs.sequenceScores.statisticalModels.differentiable.CyclicMarkovModelDiffSM
-
- setParameters(double[], int) -
Method in class de.jstacs.sequenceScores.statisticalModels.differentiable.directedGraphicalModels.BayesianNetworkDiffSM
-
- setParameters(double[], int) -
Method in class de.jstacs.sequenceScores.statisticalModels.differentiable.homogeneous.HomogeneousMM0DiffSM
-
- setParameters(double[], int) -
Method in class de.jstacs.sequenceScores.statisticalModels.differentiable.homogeneous.HomogeneousMMDiffSM
-
- setParameters(double[], int) -
Method in class de.jstacs.sequenceScores.statisticalModels.differentiable.homogeneous.UniformHomogeneousDiffSM
-
- setParameters(double[], int) -
Method in class de.jstacs.sequenceScores.statisticalModels.differentiable.IndependentProductDiffSM
-
- setParameters(double[], int) -
Method in class de.jstacs.sequenceScores.statisticalModels.differentiable.MappingDiffSM
-
- setParameters(double[], int) -
Method in class de.jstacs.sequenceScores.statisticalModels.differentiable.MarkovRandomFieldDiffSM
-
- setParameters(double[], int) -
Method in class de.jstacs.sequenceScores.statisticalModels.differentiable.mixture.AbstractMixtureDiffSM
-
- setParameters(double[], int) -
Method in class de.jstacs.sequenceScores.statisticalModels.differentiable.mixture.motif.MixtureDurationDiffSM
-
- setParameters(double[], int) -
Method in class de.jstacs.sequenceScores.statisticalModels.differentiable.mixture.motif.SkewNormalLikeDurationDiffSM
-
- setParameters(double, double, double) -
Method in class de.jstacs.sequenceScores.statisticalModels.differentiable.mixture.motif.SkewNormalLikeDurationDiffSM
- this method can be used to set the parameters even if the parameters are not allowed to be optimized.
- setParameters(double[], int) -
Method in class de.jstacs.sequenceScores.statisticalModels.differentiable.mixture.motif.UniformDurationDiffSM
-
- setParameters(double[], int) -
Method in class de.jstacs.sequenceScores.statisticalModels.differentiable.NormalizedDiffSM
-
- setParameters(double[], int) -
Method in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.models.DifferentiableHigherOrderHMM
-
- setParameters(Emission) -
Method in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.states.emissions.continuous.GaussianEmission
-
- setParameters(Emission) -
Method in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.states.emissions.continuous.MultivariateGaussianEmission
-
- setParameters(Emission) -
Method in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.states.emissions.discrete.AbstractConditionalDiscreteEmission
-
- setParameters(Emission) -
Method in interface de.jstacs.sequenceScores.statisticalModels.trainable.hmm.states.emissions.Emission
- Set values of parameters of the instance to the value of the parameters of the given instance.
- setParameters(Emission) -
Method in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.states.emissions.MixtureEmission
-
- setParameters(Emission) -
Method in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.states.emissions.SilentEmission
-
- setParameters(Emission) -
Method in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.states.emissions.UniformEmission
-
- setParameters(BasicHigherOrderTransition.AbstractTransitionElement) -
Method in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.transitions.BasicHigherOrderTransition.AbstractTransitionElement
- Set values of parameters of the instance to the value of the parameters of the given instance.
- setParameters(Transition) -
Method in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.transitions.BasicHigherOrderTransition
-
- setParameters(double[], int) -
Method in interface de.jstacs.sequenceScores.statisticalModels.trainable.hmm.transitions.DifferentiableTransition
- This method allows to set the parameters of the transition.
- setParameters(double[], int) -
Method in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.transitions.elements.TransitionElement
- This method sets the internal parameters to the values of
params beginning at index start.
- setParameters(double[], int) -
Method in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.transitions.HigherOrderTransition
-
- setParameters(Transition) -
Method in interface de.jstacs.sequenceScores.statisticalModels.trainable.hmm.transitions.Transition
- Set values of parameters of the instance to the value of the parameters of the given instance.
- setParametersForFunction(int, double[], int) -
Method in class de.jstacs.sequenceScores.statisticalModels.differentiable.mixture.AbstractMixtureDiffSM
- This method allows to set the parameters for specific functions.
- setParametersToValue(MEMConstraint[], double) -
Static method in class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.inhomogeneous.MEMTools
- This method is a convenience method that sets the same value for all parameter of the constraints
- setParams(double[]) -
Method in class de.jstacs.classifiers.differentiableSequenceScoreBased.AbstractMultiThreadedOptimizableFunction
-
- setParams(int) -
Method in class de.jstacs.classifiers.differentiableSequenceScoreBased.AbstractMultiThreadedOptimizableFunction
- This method sets the parameters for thread
index
- setParams(int) -
Method in class de.jstacs.classifiers.differentiableSequenceScoreBased.DiffSSBasedOptimizableFunction
-
- setParams(double[]) -
Method in class de.jstacs.classifiers.differentiableSequenceScoreBased.OptimizableFunction
- Sets the current values as parameters.
- setParams(double[], int) -
Method in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.transitions.HigherOrderTransition
- This method allows to set the new parameters using a specific offset.
- setParamsStarts() -
Method in class de.jstacs.sequenceScores.differentiable.IndependentProductDiffSS
- This method set the value of the array
IndependentProductDiffSS.startIndexOfParams.
- setParent(ParameterSet) -
Method in class de.jstacs.parameters.Parameter
- Sets the reference of the enclosing
ParameterSet of this
Parameter to parent.
- setParent(ParameterSetContainer) -
Method in class de.jstacs.parameters.ParameterSet
- Sets the enclosing
ParameterSetContainer of this
ParameterSet to parent.
- setParser(SequenceAnnotationParser) -
Method in class de.jstacs.results.DataSetResult
- Sets the
SequenceAnnotationParser that can be used to
write this DataSetResult including annotations on the contained Sequences
to a file
- setPath(String) -
Method in class de.jstacs.utils.galaxy.GalaxyAdaptor.FileResult
- Sets the path of the directory containing the file to
path
- setPlugInParameters(int, boolean, DataSet[], double[][]) -
Method in class de.jstacs.sequenceScores.statisticalModels.differentiable.directedGraphicalModels.BayesianNetworkDiffSM
- Computes and sets the plug-in parameters (MAP estimated parameters) from
data using weights.
- setPrior(LogPrior) -
Method in class de.jstacs.classifiers.differentiableSequenceScoreBased.gendismix.GenDisMixClassifier
- This method set a new prior that should be used for optimization.
- setRangeable(boolean) -
Method in class de.jstacs.parameters.AbstractSelectionParameter
- Sets the value returned by
AbstractSelectionParameter.isRangeable() to
rangeable.
- setRangeable(boolean) -
Method in class de.jstacs.parameters.SimpleParameter
- Sets the value returned by
SimpleParameter.isRangeable() to
rangeable.
- setRootValue(int, double) -
Method in class de.jstacs.algorithms.graphs.tensor.AsymmetricTensor
-
- setRootValue(int, double) -
Method in class de.jstacs.algorithms.graphs.tensor.SubTensor
-
- setRootValue(int, double) -
Method in class de.jstacs.algorithms.graphs.tensor.SymmetricTensor
-
- setRootValue(int, double) -
Method in class de.jstacs.algorithms.graphs.tensor.Tensor
- Sets the value
val for the root node child.
- setSeed(long) -
Method in class de.jstacs.utils.random.RandomNumberGenerator
-
- setSelected(String, boolean) -
Method in class de.jstacs.parameters.MultiSelectionParameter
- Sets the selection of the option with key
key to the value
of selected.
- setSelected(int, boolean) -
Method in class de.jstacs.parameters.MultiSelectionParameter
- Sets the selection of option with no.
- setShape(String) -
Method in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.states.emissions.discrete.AbstractConditionalDiscreteEmission
- Sets the graphviz shape of the node that uses this emission to some non-standard value
(standard is "house").
- setShiftCorrection(boolean) -
Method in class de.jstacs.sequenceScores.statisticalModels.trainable.mixture.motif.ZOOPSTrainSM
- Enables or disables the phase shift correction.
- setStartParamsToConditionalStationaryDistributions() -
Method in class de.jstacs.sequenceScores.statisticalModels.differentiable.homogeneous.HomogeneousMMDiffSM
- Sets the start parameters of this homogeneous Markov model to
the corresponding stationary distributions of the transition probabilities.
- setStatisticForHyperparameters(int[], double[]) -
Method in class de.jstacs.sequenceScores.statisticalModels.differentiable.CyclicMarkovModelDiffSM
-
- setStatisticForHyperparameters(int[], double[]) -
Method in class de.jstacs.sequenceScores.statisticalModels.differentiable.homogeneous.HomogeneousMM0DiffSM
-
- setStatisticForHyperparameters(int[], double[]) -
Method in class de.jstacs.sequenceScores.statisticalModels.differentiable.homogeneous.HomogeneousMMDiffSM
-
- setStatisticForHyperparameters(int[], double[]) -
Method in class de.jstacs.sequenceScores.statisticalModels.differentiable.homogeneous.UniformHomogeneousDiffSM
-
- setStatisticForHyperparameters(int[], double[]) -
Method in class de.jstacs.sequenceScores.statisticalModels.differentiable.mixture.VariableLengthMixtureDiffSM
-
- setStatisticForHyperparameters(int[], double[]) -
Method in interface de.jstacs.sequenceScores.statisticalModels.differentiable.VariableLengthDiffSM
- This method sets the hyperparameters for the model parameters by
evaluating the given statistic.
- setStoreAll(boolean) -
Method in class de.jstacs.classifiers.assessment.ClassifierAssessmentAssessParameterSet
- This method allows to set the switch for storing all individual performance measure values of each iteration of the
ClassifierAssessment.
- setStringToBeParsed(String) -
Method in class de.jstacs.io.SymbolExtractor
- Sets a new
String to be parsed.
- setTempDir(File) -
Method in class de.jstacs.classifiers.differentiableSequenceScoreBased.sampling.SamplingScoreBasedClassifier
- Sets the directory for parameter files set in this
SamplingScoreBasedClassifier.
- setThreadIndependentParameters() -
Method in class de.jstacs.classifiers.differentiableSequenceScoreBased.AbstractMultiThreadedOptimizableFunction
- This method allows to set thread independent parameters.
- setThreadIndependentParameters() -
Method in class de.jstacs.classifiers.differentiableSequenceScoreBased.DiffSSBasedOptimizableFunction
-
- setThresholdClassWeights(boolean, double) -
Method in class de.jstacs.classifiers.AbstractScoreBasedClassifier
- Sets a new threshold for 2-class-classifiers.
Only available if this AbstractScoreBasedClassifier distinguishes
between 2 classes 0 and 1.
- setTrainData(DataSet) -
Method in class de.jstacs.sequenceScores.statisticalModels.trainable.mixture.AbstractMixtureTrainSM
- This method is invoked by the
train-method and sets for a
given data set the data set that should be used for train.
- setTrainData(DataSet) -
Method in class de.jstacs.sequenceScores.statisticalModels.trainable.mixture.MixtureTrainSM
-
- setTrainData(DataSet) -
Method in class de.jstacs.sequenceScores.statisticalModels.trainable.mixture.motif.ZOOPSTrainSM
-
- setTrainData(DataSet) -
Method in class de.jstacs.sequenceScores.statisticalModels.trainable.mixture.StrandTrainSM
-
- setValidator(ParameterValidator) -
Method in class de.jstacs.parameters.SimpleParameter
- Sets a new
ParameterValidator for this SimpleParameter.
- setValue(byte, double, int, int...) -
Method in class de.jstacs.algorithms.graphs.tensor.AsymmetricTensor
-
- setValue(byte, double, int, int...) -
Method in class de.jstacs.algorithms.graphs.tensor.SubTensor
-
- setValue(byte, double, int, int...) -
Method in class de.jstacs.algorithms.graphs.tensor.SymmetricTensor
- Sets the value if it is bigger than the current value and keeps the
parents information.
- setValue(byte, double, int, int...) -
Method in class de.jstacs.algorithms.graphs.tensor.Tensor
- Sets the value for the edge
parents[0],...,parents[k-1] -> child.
- setValue(Object) -
Method in class de.jstacs.parameters.EnumParameter
-
- setValue(Object) -
Method in class de.jstacs.parameters.FileParameter
-
- setValue(Object) -
Method in class de.jstacs.parameters.MultiSelectionParameter
-
- setValue(Object) -
Method in class de.jstacs.parameters.Parameter
- Sets the value of this
Parameter to value.
- setValue(Object) -
Method in class de.jstacs.parameters.ParameterSetContainer
-
- setValue(Object) -
Method in class de.jstacs.parameters.RangeParameter
-
- setValue(Object) -
Method in class de.jstacs.parameters.SelectionParameter
- Sets the selected value to the one that is specified by the key
value.
- setValue(Object) -
Method in class de.jstacs.parameters.SimpleParameter
-
- setValue(double) -
Method in class de.jstacs.sampling.AbstractBurnInTest
-
- setValue(double) -
Method in interface de.jstacs.sampling.BurnInTest
- This method can be used to fill the internal memory with the values that
will be used to determine the length of the burn-in phase.
- setValue(double) -
Method in class de.jstacs.sampling.SimpleBurnInTest
- Deprecated.
- setValue(double) -
Method in class de.jstacs.sequenceScores.statisticalModels.differentiable.directedGraphicalModels.BNDiffSMParameter
- Sets the current value of this parameter.
- setValue(int) -
Method in class de.jstacs.utils.DefaultProgressUpdater
-
- setValue(int) -
Method in class de.jstacs.utils.GUIProgressUpdater
-
- setValue(int) -
Method in class de.jstacs.utils.NullProgressUpdater
-
- setValue(int) -
Method in interface de.jstacs.utils.ProgressUpdater
- Sets the current value the supervised process has reached.
- setValue(int) -
Method in class de.jstacs.utils.TimeLimitedProgressUpdater
-
- setValueFromTag(String, Object) -
Method in class de.jstacs.parameters.ParameterSetTagger
- This method allows to easily set the value of a parameter defined by the tag.
- setValues(String) -
Method in class de.jstacs.parameters.RangeParameter
- Sets a list of values from a
String containing a space separated
list of values.
- setValues(Object, int, Object, RangeParameter.Scale) -
Method in class de.jstacs.parameters.RangeParameter
- Sets the values of this
RangeParameter as a range of values,
specified by a start value, a last value, a number of steps between these
values (without the last value) and a scale in that the values between
the first and the last value are chosen.
- setValues(double[]) -
Method in class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.inhomogeneous.MEMTools.DualFunction
- This method set the values of the Lagrange multiplicators of the constraints
- setValuesInLogScale(boolean, double, Object, int, Object) -
Method in class de.jstacs.parameters.RangeParameter
- This method enables you to set a list of values in an easy manner.
- setWeight(double) -
Method in class de.jstacs.sequenceScores.statisticalModels.trainable.phylo.PhyloNode
- This method set the weight (length, rate ...) for the incoming edge
- setWeights(double...) -
Method in class de.jstacs.classifiers.differentiableSequenceScoreBased.gendismix.GenDisMixClassifier
- This method set the weights for the summand of the function.
- setWeights(double...) -
Method in class de.jstacs.sequenceScores.statisticalModels.trainable.mixture.AbstractMixtureTrainSM
- Sets the weights of each component.
- SGIS -
Static variable in class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.inhomogeneous.MEMTools
- This constant can be used to specify that the model should use the iterative scaling for
training.
- SGIS_P -
Static variable in class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.inhomogeneous.MEMTools
- This constant can be used to specify that the model should use the iterative scaling for
training.
- shallBeRanged() -
Method in class de.jstacs.parameters.RangeParameter
- Returns one of
LIST, RANGE or NO depending on
the input used to specify this RangeParameter.
- SharedStructureClassifier - Class in de.jstacs.sequenceScores.statisticalModels.trainable.discrete.inhomogeneous.shared
- This class enables you to learn the structure on all classes of the
classifier together.
- SharedStructureClassifier(int, StructureLearner.ModelType, byte, StructureLearner.LearningType, FSDAGTrainSM...) -
Constructor for class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.inhomogeneous.shared.SharedStructureClassifier
- Creates a new
SharedStructureClassifier from given
FSDAGTrainSMs.
- SharedStructureClassifier(StringBuffer) -
Constructor for class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.inhomogeneous.shared.SharedStructureClassifier
- The standard constructor for the interface
Storable.
- SharedStructureMixture - Class in de.jstacs.sequenceScores.statisticalModels.trainable.discrete.inhomogeneous.shared
- This class handles a mixture of models with the same structure that is
learned via EM.
- SharedStructureMixture(FSDAGTrainSM[], StructureLearner.ModelType, byte, int, double, TerminationCondition) -
Constructor for class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.inhomogeneous.shared.SharedStructureMixture
- Creates a new
SharedStructureMixture instance which estimates the
component probabilities/weights.
- SharedStructureMixture(FSDAGTrainSM[], StructureLearner.ModelType, byte, int, double[], double, TerminationCondition) -
Constructor for class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.inhomogeneous.shared.SharedStructureMixture
- Creates a new
SharedStructureMixture instance with fixed
component weights.
- SharedStructureMixture(FSDAGTrainSM[], StructureLearner.ModelType, byte, int, boolean, double[], double, TerminationCondition) -
Constructor for class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.inhomogeneous.shared.SharedStructureMixture
- Creates a new
SharedStructureMixture instance with all relevant
values.
- SharedStructureMixture(StringBuffer) -
Constructor for class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.inhomogeneous.shared.SharedStructureMixture
- The standard constructor for the interface
Storable.
- shortcut -
Variable in class de.jstacs.classifiers.differentiableSequenceScoreBased.DiffSSBasedOptimizableFunction
- These shortcuts indicate the beginning of a new part in the parameter vector.
- ShortSequence - Class in de.jstacs.data.sequences
- This class is for sequences with the alphabet symbols encoded as
shortss and can therefore be used for discrete
AlphabetContainers with alphabets that use many different symbols. - ShortSequence(AlphabetContainer, short[]) -
Constructor for class de.jstacs.data.sequences.ShortSequence
- Creates a new
ShortSequence from an array of short-
encoded alphabet symbols.
- ShortSequence(AlphabetContainer, String) -
Constructor for class de.jstacs.data.sequences.ShortSequence
- Creates a new
ShortSequence from a String representation
using the default delimiter.
- ShortSequence(AlphabetContainer, SequenceAnnotation[], String, String) -
Constructor for class de.jstacs.data.sequences.ShortSequence
- Creates a new
ShortSequence from a String representation
using the delimiter delim.
- ShortSequence(AlphabetContainer, SequenceAnnotation[], SymbolExtractor) -
Constructor for class de.jstacs.data.sequences.ShortSequence
- Creates a new
ShortSequence from a SymbolExtractor.
- shouldBeNormalized() -
Method in class de.jstacs.classifiers.differentiableSequenceScoreBased.gendismix.GenDisMixClassifierParameterSet
- This method indicates if a normalization shall be used while
optimization.
- showImage(String, BufferedImage) -
Static method in class de.jstacs.utils.REnvironment
- Enables you to show an image.
- showImage(String, BufferedImage, int) -
Static method in class de.jstacs.utils.REnvironment
- Enables you to show an image.
- SignificantMotifOccurrencesFinder - Class in de.jstacs.motifDiscovery
- This class enables the user to predict motif occurrences given a specific significance level.
- SignificantMotifOccurrencesFinder(MotifDiscoverer, SignificantMotifOccurrencesFinder.RandomSeqType, boolean, int, double) -
Constructor for class de.jstacs.motifDiscovery.SignificantMotifOccurrencesFinder
- This constructor creates an instance of
SignificantMotifOccurrencesFinder that uses the given SignificantMotifOccurrencesFinder.RandomSeqType to determine the siginificance level.
- SignificantMotifOccurrencesFinder(MotifDiscoverer, SignificantMotifOccurrencesFinder.RandomSeqType, SignificantMotifOccurrencesFinder.JoinMethod, boolean, int, double) -
Constructor for class de.jstacs.motifDiscovery.SignificantMotifOccurrencesFinder
- This constructor creates an instance of
SignificantMotifOccurrencesFinder that uses the given SignificantMotifOccurrencesFinder.RandomSeqType to determine the siginificance level.
- SignificantMotifOccurrencesFinder(MotifDiscoverer, DataSet, double[], double) -
Constructor for class de.jstacs.motifDiscovery.SignificantMotifOccurrencesFinder
- This constructor creates an instance of
SignificantMotifOccurrencesFinder that uses a DataSet to determine the siginificance level.
- SignificantMotifOccurrencesFinder(MotifDiscoverer, SignificantMotifOccurrencesFinder.JoinMethod, DataSet, double[], double) -
Constructor for class de.jstacs.motifDiscovery.SignificantMotifOccurrencesFinder
- This constructor creates an instance of
SignificantMotifOccurrencesFinder that uses a DataSet to determine the siginificance level.
- SignificantMotifOccurrencesFinder.JoinMethod - Interface in de.jstacs.motifDiscovery
- Interface for methods that combine several profiles over the same sequence
into one common profile
- SignificantMotifOccurrencesFinder.RandomSeqType - Enum in de.jstacs.motifDiscovery
-
- SignificantMotifOccurrencesFinder.SumOfProbabilities - Class in de.jstacs.motifDiscovery
- Joins several profiles containing log-probabilities into one profile containing
the logarithm of the sum of the probabilities of the single profiles.
- SignificantMotifOccurrencesFinder.SumOfProbabilities() -
Constructor for class de.jstacs.motifDiscovery.SignificantMotifOccurrencesFinder.SumOfProbabilities
-
- SilentEmission - Class in de.jstacs.sequenceScores.statisticalModels.trainable.hmm.states.emissions
- This class implements a silent emission which is used to create silent states.
- SilentEmission() -
Constructor for class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.states.emissions.SilentEmission
- The main constructor.
- SilentEmission(StringBuffer) -
Constructor for class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.states.emissions.SilentEmission
- The standard constructor for the interface
Storable.
- SimpleBurnInTest - Class in de.jstacs.sampling
- Deprecated. since this burn test ignore the data coming from the sampling, it might be problematic to use this test
- SimpleBurnInTest(int) -
Constructor for class de.jstacs.sampling.SimpleBurnInTest
- Deprecated. This is the main constructor that creates an instance of
SimpleBurnInTest with fixed burn-in length.
- SimpleBurnInTest(StringBuffer) -
Constructor for class de.jstacs.sampling.SimpleBurnInTest
- Deprecated. The standard constructor for the interface
Storable.
- SimpleCosts - Class in de.jstacs.algorithms.alignment.cost
- Class for simple costs with costs
match for a match,
mismatch for a mismatch, and
gap for a gap (of length 1).
- SimpleCosts(double, double, double) -
Constructor for class de.jstacs.algorithms.alignment.cost.SimpleCosts
- Creates a new instance of simple costs with costs
match for a match,
mismatch for a mismatch, and
gap for a gap (of length 1).
- SimpleDifferentiableState - Class in de.jstacs.sequenceScores.statisticalModels.trainable.hmm.states
- This class implements a
State based on Emission that allows to reuse Emissions for different States. - SimpleDifferentiableState(DifferentiableEmission, String, boolean) -
Constructor for class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.states.SimpleDifferentiableState
- This is the constructor of a
SimpleState.
- SimpleDiscreteSequence - Class in de.jstacs.data.sequences
- This is the main class for any discrete sequence.
- SimpleDiscreteSequence(AlphabetContainer, SequenceAnnotation[]) -
Constructor for class de.jstacs.data.sequences.SimpleDiscreteSequence
- This constructor creates a new
SimpleDiscreteSequence with the
AlphabetContainer container and the annotation
annotation but without the content.
- SimpleGaussianSumLogPrior - Class in de.jstacs.classifiers.differentiableSequenceScoreBased.logPrior
- This class implements a prior that is a product of Gaussian distributions
with mean 0 and equal variance for each parameter.
- SimpleGaussianSumLogPrior(double) -
Constructor for class de.jstacs.classifiers.differentiableSequenceScoreBased.logPrior.SimpleGaussianSumLogPrior
- Creates a new
SimpleGaussianSumLogPrior with mean 0 and variance
sigma for all parameters, including the class parameters.
- SimpleGaussianSumLogPrior(StringBuffer) -
Constructor for class de.jstacs.classifiers.differentiableSequenceScoreBased.logPrior.SimpleGaussianSumLogPrior
- The standard constructor for the interface
Storable.
- SimpleHistory - Class in de.jstacs.motifDiscovery.history
- This class implements a simple history that has a limited memory that will be
used cyclicly.
- SimpleHistory(int) -
Constructor for class de.jstacs.motifDiscovery.history.SimpleHistory
- This constructor creates a simple history with limited memory.
- SimpleHistory(int, boolean, boolean, boolean) -
Constructor for class de.jstacs.motifDiscovery.history.SimpleHistory
- This constructor creates a simple history with limited memory.
- SimpleHistory(StringBuffer) -
Constructor for class de.jstacs.motifDiscovery.history.SimpleHistory
- This is the constructor for the interface
Storable.
- SimpleParameter - Class in de.jstacs.parameters
- Class for a "simple" parameter.
- SimpleParameter(StringBuffer) -
Constructor for class de.jstacs.parameters.SimpleParameter
- The standard constructor for the interface
Storable.
- SimpleParameter(DataType, String, String, boolean) -
Constructor for class de.jstacs.parameters.SimpleParameter
- Constructor for a
SimpleParameter without
ParameterValidator.
- SimpleParameter(DataType, String, String, boolean, Object) -
Constructor for class de.jstacs.parameters.SimpleParameter
- Constructor for a
SimpleParameter without
ParameterValidator but with a default value.
- SimpleParameter(DataType, String, String, boolean, ParameterValidator) -
Constructor for class de.jstacs.parameters.SimpleParameter
- Constructor for a
SimpleParameter with a
ParameterValidator.
- SimpleParameter(DataType, String, String, boolean, ParameterValidator, Object) -
Constructor for class de.jstacs.parameters.SimpleParameter
- Constructor for a
SimpleParameter with validator and default
value.
- SimpleParameter.DatatypeNotValidException - Exception in de.jstacs.parameters
- Class for an
Exception that can be thrown if the provided
int-value that represents a data type is not one of the
values defined in DataType. - SimpleParameter.DatatypeNotValidException(String) -
Constructor for exception de.jstacs.parameters.SimpleParameter.DatatypeNotValidException
- Creates a new
SimpleParameter.DatatypeNotValidException with an error
message.
- SimpleParameter.IllegalValueException - Exception in de.jstacs.parameters
- This exception is thrown if a parameter is not valid.
- SimpleParameter.IllegalValueException(String) -
Constructor for exception de.jstacs.parameters.SimpleParameter.IllegalValueException
- Creates a new
SimpleParameter.IllegalValueException with the reason of the
exception reason as error message.
- SimpleParameterSet - Class in de.jstacs.parameters
- Class for a
ParameterSet that is constructed from an array of Parameters. - SimpleParameterSet(Parameter...) -
Constructor for class de.jstacs.parameters.SimpleParameterSet
- Creates a new
SimpleParameterSet from an array of Parameters.
- SimpleParameterSet(StringBuffer) -
Constructor for class de.jstacs.parameters.SimpleParameterSet
- The standard constructor for the interface
Storable.
- SimpleResult - Class in de.jstacs.results
- Abstract class for a
Result with a value of a primitive data type or
String. - SimpleResult(String, String, DataType) -
Constructor for class de.jstacs.results.SimpleResult
- The main constructor which takes the main information of a result.
- SimpleResult(StringBuffer) -
Constructor for class de.jstacs.results.SimpleResult
- This is the constructor for
Storable.
- SimpleSamplingState - Class in de.jstacs.sequenceScores.statisticalModels.trainable.hmm.states
- This class implements a state that can be used for a HMM that obtains its parameters from sampling.
- SimpleSamplingState(SamplingEmission, String, boolean) -
Constructor for class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.states.SimpleSamplingState
- This constructor creates a state that can be used in a HMM that obtains its parameters from sampling.
- SimpleSequenceAnnotationParser - Class in de.jstacs.data.sequences.annotation
- This class implements a naive
SequenceAnnotationParser which simply paste the comments into SequenceAnnotation. - SimpleSequenceAnnotationParser() -
Constructor for class de.jstacs.data.sequences.annotation.SimpleSequenceAnnotationParser
- The constructor of a
SimpleSequenceAnnotationParser which simply paste the comments into SequenceAnnotation.
- SimpleSequenceIterator - Class in de.jstacs.data.bioJava
- Class that implements the
SequenceIterator interface of BioJava in a
simple way, backed by an array of Sequences. - SimpleSequenceIterator(Sequence...) -
Constructor for class de.jstacs.data.bioJava.SimpleSequenceIterator
- Creates a new
SimpleSequenceIterator from an array of
Sequences.
- SimpleState - Class in de.jstacs.sequenceScores.statisticalModels.trainable.hmm.states
- This class implements a
State based on Emission that allows to reuse Emissions for different States. - SimpleState(Emission, String, boolean) -
Constructor for class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.states.SimpleState
- This is the constructor of a
SimpleState.
- SimpleStaticConstraint - Class in de.jstacs.parameters.validation
- Class for a
Constraint that checks values against static values using
the comparison operators defined in the interface Constraint. - SimpleStaticConstraint(Number, int) -
Constructor for class de.jstacs.parameters.validation.SimpleStaticConstraint
- Creates a new
SimpleStaticConstraint from a Number
-reference and a comparison operator as defined in Constraint.
- SimpleStaticConstraint(String, int) -
Constructor for class de.jstacs.parameters.validation.SimpleStaticConstraint
- Creates a new
SimpleStaticConstraint from a String
-reference and a comparison operator as defined in Constraint.
- SimpleStaticConstraint(StringBuffer) -
Constructor for class de.jstacs.parameters.validation.SimpleStaticConstraint
- The standard constructor for the interface
Storable.
- SimpleStringExtractor - Class in de.jstacs.io
- This is a simple class that extracts
Strings. - SimpleStringExtractor(String...) -
Constructor for class de.jstacs.io.SimpleStringExtractor
- This constructor packs the
Strings in an instance of
SimpleStringExtractor.
- simpleWeights(double[]) -
Static method in class de.jstacs.classifiers.performanceMeasures.AbstractPerformanceMeasure
- Returns true if all weights in
weight are 1.
- SinglePositionSequenceAnnotation - Class in de.jstacs.data.sequences.annotation
- Class for some annotations that consist mainly of one position on a sequence.
- SinglePositionSequenceAnnotation(SinglePositionSequenceAnnotation.Type, String, int) -
Constructor for class de.jstacs.data.sequences.annotation.SinglePositionSequenceAnnotation
- Creates a new
SinglePositionSequenceAnnotation of type
type with identifier identifier and position
position.
- SinglePositionSequenceAnnotation(SinglePositionSequenceAnnotation.Type, String, int, Result...) -
Constructor for class de.jstacs.data.sequences.annotation.SinglePositionSequenceAnnotation
- Creates a new
SinglePositionSequenceAnnotation of type
type with identifier identifier, position
position and additional annotations
additionalAnnotation.
- SinglePositionSequenceAnnotation(StringBuffer) -
Constructor for class de.jstacs.data.sequences.annotation.SinglePositionSequenceAnnotation
- The standard constructor for the interface
Storable.
- SinglePositionSequenceAnnotation.Type - Enum in de.jstacs.data.sequences.annotation
- This
enum defines possible types of a
SinglePositionSequenceAnnotation. - SINGLETON -
Static variable in class de.jstacs.data.alphabets.DNAAlphabet.DNAAlphabetParameterSet
- The only instance of this class.
- SINGLETON -
Static variable in class de.jstacs.data.alphabets.DNAAlphabet
- The only instance of this class.
- SINGLETON -
Static variable in class de.jstacs.data.alphabets.DNAAlphabetContainer.DNAAlphabetContainerParameterSet
- The only instance of this class.
- SINGLETON -
Static variable in class de.jstacs.data.alphabets.DNAAlphabetContainer
- The only instance of this class.
- SINGLETON -
Static variable in class de.jstacs.data.alphabets.ProteinAlphabet.ProteinAlphabetParameterSet
- The only instance of this class.
- SINGLETON -
Static variable in class de.jstacs.data.alphabets.ProteinAlphabet
- The only instance of this class.
- Singleton - Interface in de.jstacs
- This interface states that the implementing class has only one immutable instance.
- Singleton.SingletonHandler - Class in de.jstacs
- This handler helps to retrieve the single instance of a
Singleton. - Singleton.SingletonHandler() -
Constructor for class de.jstacs.Singleton.SingletonHandler
-
- size() -
Method in class de.jstacs.AnnotatedEntityList
- Returns the number of
AnnotatedEntitys (not the capacity)
in the AnnotatedEntityList.
- SkewNormalLikeDurationDiffSM - Class in de.jstacs.sequenceScores.statisticalModels.differentiable.mixture.motif
- This class implements a skew normal like discrete truncated distribution.
- SkewNormalLikeDurationDiffSM(int, int, double, double, double) -
Constructor for class de.jstacs.sequenceScores.statisticalModels.differentiable.mixture.motif.SkewNormalLikeDurationDiffSM
- This is the main constructor if the parameters are fixed.
- SkewNormalLikeDurationDiffSM(int, int, boolean, double, double, boolean, double, double, boolean, double, double, int) -
Constructor for class de.jstacs.sequenceScores.statisticalModels.differentiable.mixture.motif.SkewNormalLikeDurationDiffSM
- This is the constructor that allows the most flexible handling of the parameters.
- SkewNormalLikeDurationDiffSM(StringBuffer) -
Constructor for class de.jstacs.sequenceScores.statisticalModels.differentiable.mixture.motif.SkewNormalLikeDurationDiffSM
- This is the constructor for
Storable.
- skip(int) -
Method in class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.inhomogeneous.SequenceIterator
- This method skips some position.
- skipInit -
Variable in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.models.HigherOrderHMM
- Indicates if the model should be initialized (randomly) before optimization
- skipLastClassifiersDuringClassifierTraining -
Variable in class de.jstacs.classifiers.assessment.ClassifierAssessment
- Skip last classifier.
- SmallDifferenceOfFunctionEvaluationsCondition - Class in de.jstacs.algorithms.optimization.termination
- This class implements a
TerminationCondition that stops an optimization
if the difference of the current and the last function evaluations will be small, i.e.,
 .
. - SmallDifferenceOfFunctionEvaluationsCondition(double) -
Constructor for class de.jstacs.algorithms.optimization.termination.SmallDifferenceOfFunctionEvaluationsCondition
- This constructor creates an instance that stops the optimization if the difference of the
current and the last function evaluations is smaller than
epsilon.
- SmallDifferenceOfFunctionEvaluationsCondition(SmallDifferenceOfFunctionEvaluationsCondition.SmallDifferenceOfFunctionEvaluationsConditionParameterSet) -
Constructor for class de.jstacs.algorithms.optimization.termination.SmallDifferenceOfFunctionEvaluationsCondition
- This is the main constructor creating an instance from a given parameter set.
- SmallDifferenceOfFunctionEvaluationsCondition(StringBuffer) -
Constructor for class de.jstacs.algorithms.optimization.termination.SmallDifferenceOfFunctionEvaluationsCondition
- The standard constructor for the interface
Storable.
- SmallDifferenceOfFunctionEvaluationsCondition.SmallDifferenceOfFunctionEvaluationsConditionParameterSet - Class in de.jstacs.algorithms.optimization.termination
- This class implements the parameter set for a
SmallDifferenceOfFunctionEvaluationsCondition. - SmallDifferenceOfFunctionEvaluationsCondition.SmallDifferenceOfFunctionEvaluationsConditionParameterSet() -
Constructor for class de.jstacs.algorithms.optimization.termination.SmallDifferenceOfFunctionEvaluationsCondition.SmallDifferenceOfFunctionEvaluationsConditionParameterSet
- This constructor creates an empty parameter set.
- SmallDifferenceOfFunctionEvaluationsCondition.SmallDifferenceOfFunctionEvaluationsConditionParameterSet(StringBuffer) -
Constructor for class de.jstacs.algorithms.optimization.termination.SmallDifferenceOfFunctionEvaluationsCondition.SmallDifferenceOfFunctionEvaluationsConditionParameterSet
- The standard constructor for the interface
Storable.
- SmallDifferenceOfFunctionEvaluationsCondition.SmallDifferenceOfFunctionEvaluationsConditionParameterSet(double) -
Constructor for class de.jstacs.algorithms.optimization.termination.SmallDifferenceOfFunctionEvaluationsCondition.SmallDifferenceOfFunctionEvaluationsConditionParameterSet
- This constructor creates a filled instance of a parameters set.
- SmallGradientConditon - Class in de.jstacs.algorithms.optimization.termination
- This class implements a
TerminationCondition that allows no further iteration in an optimization if the
the gradient becomes small, i.e.,
 .
. - SmallGradientConditon(double) -
Constructor for class de.jstacs.algorithms.optimization.termination.SmallGradientConditon
- This constructor creates an instance that stops the optimization if the sum of the absolute
values of gradient components is smaller than
epsilon.
- SmallGradientConditon(SmallGradientConditon.SmallGradientConditonParameterSet) -
Constructor for class de.jstacs.algorithms.optimization.termination.SmallGradientConditon
- This is the main constructor creating an instance from a given parameter set.
- SmallGradientConditon(StringBuffer) -
Constructor for class de.jstacs.algorithms.optimization.termination.SmallGradientConditon
- The standard constructor for the interface
Storable.
- SmallGradientConditon.SmallGradientConditonParameterSet - Class in de.jstacs.algorithms.optimization.termination
- This class implements the parameter set for a
SmallStepCondition. - SmallGradientConditon.SmallGradientConditonParameterSet() -
Constructor for class de.jstacs.algorithms.optimization.termination.SmallGradientConditon.SmallGradientConditonParameterSet
- This constructor creates an empty parameter set.
- SmallGradientConditon.SmallGradientConditonParameterSet(StringBuffer) -
Constructor for class de.jstacs.algorithms.optimization.termination.SmallGradientConditon.SmallGradientConditonParameterSet
- The standard constructor for the interface
Storable.
- SmallGradientConditon.SmallGradientConditonParameterSet(double) -
Constructor for class de.jstacs.algorithms.optimization.termination.SmallGradientConditon.SmallGradientConditonParameterSet
- This constructor creates a filled instance of a parameters set.
- SmallStepCondition - Class in de.jstacs.algorithms.optimization.termination
- This class implements a
TerminationCondition that allows no further iteration in an optimization if the
scalar product of the current and the last values of x will be small, i.e.,
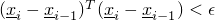 .
. - SmallStepCondition(double) -
Constructor for class de.jstacs.algorithms.optimization.termination.SmallStepCondition
- This constructor creates an instance that allows no further iteration in an optimization if the
scalar product of the current and the last values of
x is smaller than epsilon.
- SmallStepCondition(SmallStepCondition.SmallStepConditionParameterSet) -
Constructor for class de.jstacs.algorithms.optimization.termination.SmallStepCondition
- This is the main constructor creating an instance from a given parameter set.
- SmallStepCondition(StringBuffer) -
Constructor for class de.jstacs.algorithms.optimization.termination.SmallStepCondition
- The standard constructor for the interface
Storable.
- SmallStepCondition.SmallStepConditionParameterSet - Class in de.jstacs.algorithms.optimization.termination
- This class implements the parameter set for a
SmallStepCondition. - SmallStepCondition.SmallStepConditionParameterSet() -
Constructor for class de.jstacs.algorithms.optimization.termination.SmallStepCondition.SmallStepConditionParameterSet
- This constructor creates an empty parameter set.
- SmallStepCondition.SmallStepConditionParameterSet(StringBuffer) -
Constructor for class de.jstacs.algorithms.optimization.termination.SmallStepCondition.SmallStepConditionParameterSet
- The standard constructor for the interface
Storable.
- SmallStepCondition.SmallStepConditionParameterSet(double) -
Constructor for class de.jstacs.algorithms.optimization.termination.SmallStepCondition.SmallStepConditionParameterSet
- This constructor creates a filled instance of a parameters set.
- smooth(double[]) -
Method in class de.jstacs.data.DinucleotideProperty.MeanSmoothing
-
- smooth(double[]) -
Method in class de.jstacs.data.DinucleotideProperty.MedianSmoothing
-
- smooth(double[]) -
Method in class de.jstacs.data.DinucleotideProperty.NoSmoothing
-
- smooth(double[]) -
Method in class de.jstacs.data.DinucleotideProperty.Smoothing
- Returns the smoothed version of
original.
- SoftOneOfN - Class in de.jstacs.utils.random
- This random generator returns
1-epsilon for one and equal parts
for the rest of a random vector. - SoftOneOfN(double) -
Constructor for class de.jstacs.utils.random.SoftOneOfN
- This constructor can be used for (soft) sampling one of n.
- SoftOneOfN() -
Constructor for class de.jstacs.utils.random.SoftOneOfN
- This constructor can be used for (hard) sampling one of n.
- sort(String) -
Method in class de.jstacs.results.ListResult
- This method enables you to sort the entries of this container by a
specified column.
- sort() -
Method in class de.jstacs.utils.IntList
- This method sorts the elements of the list.
- sortAlongWith(double[], double[]...) -
Static method in class de.jstacs.utils.ToolBox
- This method implements a sort algorithm on the array
arrayToBeSorted.
- sostream -
Variable in class de.jstacs.classifiers.differentiableSequenceScoreBased.ScoreClassifier
- This stream is used for comments, e.g.
- sostream -
Variable in class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.inhomogeneous.InhomogeneousDGTrainSM
- This stream is used for comments, computation steps/results or any other
kind of output during the training, ...
- sostream -
Variable in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.AbstractHMM
- This is the stream for writing information while training.
- sostream -
Variable in class de.jstacs.sequenceScores.statisticalModels.trainable.mixture.AbstractMixtureTrainSM
- This is the stream for writing information while training.
- source -
Variable in class de.jstacs.algorithms.graphs.Edge
- The source node.
- SparseSequence - Class in de.jstacs.data.sequences
- This class is an implementation for sequences on one alphabet with length 4.
- SparseSequence(AlphabetContainer, String) -
Constructor for class de.jstacs.data.sequences.SparseSequence
- Creates a new
SparseSequence from a String
representation.
- SparseSequence(AlphabetContainer, SymbolExtractor) -
Constructor for class de.jstacs.data.sequences.SparseSequence
- Creates a new
SparseSequence from a SymbolExtractor.
- SparseStringExtractor - Class in de.jstacs.io
- This
StringExtractor reads the Strings from a File as
the user asks for the String. - SparseStringExtractor(String) -
Constructor for class de.jstacs.io.SparseStringExtractor
- A constructor that reads the lines from a file.
- SparseStringExtractor(File) -
Constructor for class de.jstacs.io.SparseStringExtractor
- A constructor that reads the lines from a file.
- SparseStringExtractor(String, SequenceAnnotationParser) -
Constructor for class de.jstacs.io.SparseStringExtractor
- A constructor that reads the lines from a file.
- SparseStringExtractor(String, char) -
Constructor for class de.jstacs.io.SparseStringExtractor
- A constructor that reads the lines from a file and ignores
those starting with the comment character
ignore.
- SparseStringExtractor(File, char) -
Constructor for class de.jstacs.io.SparseStringExtractor
- A constructor that reads the lines from a file and ignores
those starting with the comment character
ignore.
- SparseStringExtractor(String, char, SequenceAnnotationParser) -
Constructor for class de.jstacs.io.SparseStringExtractor
- A constructor that reads the lines from a file and ignores
those starting with the comment character
ignore.
- SparseStringExtractor(String, String, SequenceAnnotationParser) -
Constructor for class de.jstacs.io.SparseStringExtractor
- A constructor that reads the lines from a file and sets the
annotation of the source to
annotation.
- SparseStringExtractor(String, char, String, SequenceAnnotationParser) -
Constructor for class de.jstacs.io.SparseStringExtractor
- A constructor that reads the lines from a file, ignores those
starting with the comment character
ignore and sets the
annotation of the source to annotation.
- SparseStringExtractor(File, char, String, SequenceAnnotationParser) -
Constructor for class de.jstacs.io.SparseStringExtractor
- A constructor that reads the lines from a file, ignores those
starting with the comment character
ignore and sets the
annotation of the source to annotation.
- SparseStringExtractor(Reader, char, String, SequenceAnnotationParser) -
Constructor for class de.jstacs.io.SparseStringExtractor
- A constructor that reads the lines from a
Reader, ignores those
starting with the comment character ignore and sets the
annotation of the source to annotation.
- spearmanCorrelation(double[], double[]) -
Static method in class de.jstacs.utils.ToolBox
- The method computes the Spearman correlation of two vectors.
- SplitSequenceAnnotationParser - Class in de.jstacs.data.sequences.annotation
- This class implements a simple
SequenceAnnotationParser which simply splits the comments by specific delimiters. - SplitSequenceAnnotationParser() -
Constructor for class de.jstacs.data.sequences.annotation.SplitSequenceAnnotationParser
- Creates a new
SplitSequenceAnnotationParser with specific delimiters, i.e., key value
delimiter "=" and annotation delimiter ";".
- SplitSequenceAnnotationParser(String, String) -
Constructor for class de.jstacs.data.sequences.annotation.SplitSequenceAnnotationParser
- Creates a new
SplitSequenceAnnotationParser with user-specified delimiters.
- standardDeviation -
Variable in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.states.emissions.continuous.PluginGaussianEmission
- Initial standard deviation.
- start -
Variable in class de.jstacs.sequenceScores.differentiable.IndependentProductDiffSS
- This array specifies the start positions of the specific parts.
- START_NODE -
Static variable in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.AbstractHMM
- The
String for the start node used in Graphviz annotation.
- STARTDISTANCE -
Static variable in class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.inhomogeneous.MEMTools
- The start distance for the line search in an optimization using the
Optimizer.
- StartDistanceForecaster - Interface in de.jstacs.algorithms.optimization
- This interface is used to determine the next start distance that will be used
in a line search.
- startIndexOfParams -
Variable in class de.jstacs.sequenceScores.differentiable.IndependentProductDiffSS
- This array contains the start indices for
DifferentiableSequenceScore.setParameters(double[], int) on IndependentProductDiffSS.score.
- starts -
Variable in class de.jstacs.sequenceScores.statisticalModels.trainable.CompositeTrainSM
- The start indices.
- starts -
Variable in class de.jstacs.sequenceScores.statisticalModels.trainable.mixture.AbstractMixtureTrainSM
- The number of starts.
- State - Interface in de.jstacs.sequenceScores.statisticalModels.trainable.hmm.states
- This interface declares the methods of any state used in a hidden Markov model.
- stateList -
Variable in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.models.HigherOrderHMM
- Helper variable = only for internal use.
- states -
Variable in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.AbstractHMM
- The (hidden) states of the HMM.
- states -
Variable in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.transitions.BasicHigherOrderTransition.AbstractTransitionElement
- The states that can be visited
- StationaryDistribution - Class in de.jstacs.utils
- This class can be used to determine the stationary distribution.
- StationaryDistribution() -
Constructor for class de.jstacs.utils.StationaryDistribution
-
- stationaryIteration -
Variable in class de.jstacs.sequenceScores.statisticalModels.trainable.mixture.AbstractMixtureTrainSM
- The number of (stationary) iterations of the Gibbs Sampler.
- statistic -
Variable in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.states.emissions.discrete.AbstractConditionalDiscreteEmission
- The array for storing the statistics for
each parameter
- statistic -
Variable in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.transitions.BasicHigherOrderTransition.AbstractTransitionElement
- The sufficient statistic for determining the parameters during sampling, viterbi or Baum-Welch training.
- StatisticalModel - Interface in de.jstacs.sequenceScores.statisticalModels
- This interface declares methods of a statistical model, i.e., a
SequenceScore that defines a proper likelihood
over the input Sequences. - StatisticalModelTester - Class in de.jstacs.utils
- This class is useful for some test for any (discrete) models.
- StatisticalModelTester() -
Constructor for class de.jstacs.utils.StatisticalModelTester
-
- StatisticalTest - Class in de.jstacs.utils
- This class enables the user to compute some divergences.
- StatisticalTest() -
Constructor for class de.jstacs.utils.StatisticalTest
-
- statisticsTransitionProb -
Variable in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.transitions.elements.DistanceBasedScaledTransitionElement
- Represents the summarized epsilons required for estimating the transition probabilities from the
context.
- statisticsTransitionProb -
Variable in class de.jstacs.sequenceScores.statisticalModels.trainable.hmm.transitions.elements.ReferenceBasedTransitionElement
- Represents the gammas required for estimating the transition probabilities not including pseudocounts.
- STEEPEST_DESCENT -
Static variable in class de.jstacs.algorithms.optimization.Optimizer
- This constant can be used to specify that the steepest descent should be
used in the
optimize-method.
- steepestDescent(DifferentiableFunction, double[], TerminationCondition, double, StartDistanceForecaster, OutputStream, Time) -
Static method in class de.jstacs.algorithms.optimization.Optimizer
- The steepest descent.
- stopThreads() -
Method in class de.jstacs.classifiers.differentiableSequenceScoreBased.AbstractMultiThreadedOptimizableFunction
- This method can and should be used to stop all threads if they are not needed any longer.
- Storable - Interface in de.jstacs
- This is the root interface for all immutable objects that must be stored in
e.g.
- StorableResult - Class in de.jstacs.results
- Class for
Results that are Storables. - StorableResult(String, String, Storable) -
Constructor for class de.jstacs.results.StorableResult
- Creates a result for an XML representation of an object.
- StorableResult(StringBuffer) -
Constructor for class de.jstacs.results.StorableResult
- The standard constructor for the interface
Storable.
- StorableValidator - Class in de.jstacs.parameters.validation
- Class for a validator that validates instances and XML representations for
the correct class types (e.g.
- StorableValidator(Class<? extends Storable>, boolean) -
Constructor for class de.jstacs.parameters.validation.StorableValidator
- Creates a new
StorableValidator for a subclass of
AbstractTrainableStatisticalModel or AbstractClassifier.
- StorableValidator(Class<? extends Storable>) -
Constructor for class de.jstacs.parameters.validation.StorableValidator
- Creates a new
StorableValidator for a subclass of
Storable.
- StorableValidator(StringBuffer) -
Constructor for class de.jstacs.parameters.validation.StorableValidator
- The standard constructor for the interface
Storable.
- StrandDiffSM - Class in de.jstacs.sequenceScores.statisticalModels.differentiable.mixture
- This class enables the user to search on both strand.
- StrandDiffSM(DifferentiableStatisticalModel, double, int, boolean, StrandDiffSM.InitMethod) -
Constructor for class de.jstacs.sequenceScores.statisticalModels.differentiable.mixture.StrandDiffSM
- This constructor creates a StrandDiffSM that optimizes the usage of each strand.
- StrandDiffSM(DifferentiableStatisticalModel, int, boolean, StrandDiffSM.InitMethod, double) -
Constructor for class de.jstacs.sequenceScores.statisticalModels.differentiable.mixture.StrandDiffSM
- This constructor creates a StrandDiffSM that has a fixed frequency for the strand usage.
- StrandDiffSM(StringBuffer) -
Constructor for class de.jstacs.sequenceScores.statisticalModels.differentiable.mixture.StrandDiffSM
- This is the constructor for
Storable.
- StrandDiffSM.InitMethod - Enum in de.jstacs.sequenceScores.statisticalModels.differentiable.mixture
- This enum defines the different types of plug-in initialization of a
StrandDiffSM. - StrandedLocatedSequenceAnnotationWithLength - Class in de.jstacs.data.sequences.annotation
- Class for a
SequenceAnnotation that has a position, a length and an
orientation on the strand of a Sequence. - StrandedLocatedSequenceAnnotationWithLength(int, int, StrandedLocatedSequenceAnnotationWithLength.Strand, String, String, Result...) -
Constructor for class de.jstacs.data.sequences.annotation.StrandedLocatedSequenceAnnotationWithLength
- Creates a new
StrandedLocatedSequenceAnnotationWithLength of type
type with identifier identifier and additional
annotation (that does not fit the SequenceAnnotation definitions)
given as an array of Results results.
- StrandedLocatedSequenceAnnotationWithLength(int, int, StrandedLocatedSequenceAnnotationWithLength.Strand, String, String, Collection<Result>) -
Constructor for class de.jstacs.data.sequences.annotation.StrandedLocatedSequenceAnnotationWithLength
- Creates a new
StrandedLocatedSequenceAnnotationWithLength of type
type with identifier identifier and additional
annotation (that does not fit the SequenceAnnotation definitions)
given as a Collection of Results results.
- StrandedLocatedSequenceAnnotationWithLength(int, int, StrandedLocatedSequenceAnnotationWithLength.Strand, String, String, SequenceAnnotation[], Result...) -
Constructor for class de.jstacs.data.sequences.annotation.StrandedLocatedSequenceAnnotationWithLength
- Creates a new
StrandedLocatedSequenceAnnotationWithLength of type
type with identifier identifier, additional
annotation (that does not fit the SequenceAnnotation definitions)
given as an array of Results additionalAnnotations
and sub-annotations annotations.
- StrandedLocatedSequenceAnnotationWithLength(String, String, StrandedLocatedSequenceAnnotationWithLength.Strand, LocatedSequenceAnnotation[], Result...) -
Constructor for class de.jstacs.data.sequences.annotation.StrandedLocatedSequenceAnnotationWithLength
- Creates a new
StrandedLocatedSequenceAnnotationWithLength of type
type with identifier identifier, additional
annotation (that does not fit the SequenceAnnotation definitions)
given as an array of Results additionalAnnotations
and sub-annotations annotations.
- StrandedLocatedSequenceAnnotationWithLength(StringBuffer) -
Constructor for class de.jstacs.data.sequences.annotation.StrandedLocatedSequenceAnnotationWithLength
- The standard constructor for the interface
Storable.
- StrandedLocatedSequenceAnnotationWithLength.Strand - Enum in de.jstacs.data.sequences.annotation
- This enum defines possible orientations on the strands.
- strandedness() -
Method in enum de.jstacs.data.sequences.annotation.StrandedLocatedSequenceAnnotationWithLength.Strand
- Returns the strandedness, i.e.
- StrandTrainSM - Class in de.jstacs.sequenceScores.statisticalModels.trainable.mixture
- This model handles sequences that can either lie on the forward strand or on
the reverse complementary strand.
- StrandTrainSM(TrainableStatisticalModel, int, boolean, double[], double, AbstractMixtureTrainSM.Algorithm, double, TerminationCondition, AbstractMixtureTrainSM.Parameterization, int, int, BurnInTest) -
Constructor for class de.jstacs.sequenceScores.statisticalModels.trainable.mixture.StrandTrainSM
- Creates a new
StrandTrainSM.
- StrandTrainSM(TrainableStatisticalModel, int, double[], double, TerminationCondition, AbstractMixtureTrainSM.Parameterization) -
Constructor for class de.jstacs.sequenceScores.statisticalModels.trainable.mixture.StrandTrainSM
- Creates an instance using EM and estimating the component probabilities.
- StrandTrainSM(TrainableStatisticalModel, int, double, double, TerminationCondition, AbstractMixtureTrainSM.Parameterization) -
Constructor for class de.jstacs.sequenceScores.statisticalModels.trainable.mixture.StrandTrainSM
- Creates an instance using EM and fixed component probabilities.
- StrandTrainSM(TrainableStatisticalModel, int, double[], int, int, BurnInTest) -
Constructor for class de.jstacs.sequenceScores.statisticalModels.trainable.mixture.StrandTrainSM
- Creates an instance using Gibbs Sampling and sampling the component
probabilities.
- StrandTrainSM(TrainableStatisticalModel, int, double, int, int, BurnInTest) -
Constructor for class de.jstacs.sequenceScores.statisticalModels.trainable.mixture.StrandTrainSM
- Creates an instance using Gibbs Sampling and fixed component
probabilities.
- StrandTrainSM(StringBuffer) -
Constructor for class de.jstacs.sequenceScores.statisticalModels.trainable.mixture.StrandTrainSM
- The constructor for the interface
Storable.
- StringAlignment - Class in de.jstacs.algorithms.alignment
- Class for the representation of an alignment of
Strings. - StringAlignment(double, String...) -
Constructor for class de.jstacs.algorithms.alignment.StringAlignment
- This constructor creates an instance storing the aligned Strings and the costs of the alignment.
- StringAlignment(double, String[], Result) -
Constructor for class de.jstacs.algorithms.alignment.StringAlignment
- This constructor creates an instance storing the aligned Strings and the costs of the alignment.
- StringExtractor - Class in de.jstacs.io
- This class implements the reader that extracts
Strings from either a
File or a String. - StringExtractor(File, int) -
Constructor for class de.jstacs.io.StringExtractor
- A constructor that reads the lines from
file.
- StringExtractor(File, int, char) -
Constructor for class de.jstacs.io.StringExtractor
- A constructor that reads the lines from
file and ignores
those starting with the comment character ignore.
- StringExtractor(File, int, String) -
Constructor for class de.jstacs.io.StringExtractor
- A constructor that reads the lines from
file and sets the
annotation of the source to annotation.
- StringExtractor(File, int, char, String) -
Constructor for class de.jstacs.io.StringExtractor
- A constructor that reads the lines from
file, ignores those
starting with the comment character ignore and sets the
annotation of the source to annotation.
- StringExtractor(String, int, String) -
Constructor for class de.jstacs.io.StringExtractor
- A constructor that reads the lines from a
String
content and sets the annotation of the source to
annotation.
- StringExtractor(String, int, char, String) -
Constructor for class de.jstacs.io.StringExtractor
- A constructor that reads the lines from a
String
content, ignores those starting with the comment character
ignore and sets the annotation of the source to
annotation.
- StructureLearner - Class in de.jstacs.sequenceScores.statisticalModels.trainable.discrete.inhomogeneous
- This class can be used to learn the structure of any discrete model.
- StructureLearner(AlphabetContainer, int, double) -
Constructor for class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.inhomogeneous.StructureLearner
- Creates a new
StructureLearner for a given
AlphabetContainer, a given length and a given equivalent
sample size (ess).
- StructureLearner(AlphabetContainer, int) -
Constructor for class de.jstacs.sequenceScores.statisticalModels.trainable.discrete.inhomogeneous.StructureLearner
- Creates a
StructureLearner with equivalent sample
size (ess) = 0.
- StructureLearner.LearningType - Enum in de.jstacs.sequenceScores.statisticalModels.trainable.discrete.inhomogeneous
- This
enum defines the different types of learning that are
possible with the StructureLearner. - StructureLearner.ModelType - Enum in de.jstacs.sequenceScores.statisticalModels.trainable.discrete.inhomogeneous
- This
enum defines the different types of models that can be
learned with the StructureLearner. - structureMeasure -
Variable in class de.jstacs.sequenceScores.statisticalModels.differentiable.directedGraphicalModels.BayesianNetworkDiffSM
Measure that defines the network structure.
- stylesheet -
Static variable in class de.jstacs.utils.galaxy.GalaxyAdaptor
- The stylesheet used for the Galaxy HTML output.
- SubclassFinder - Class in de.jstacs.utils
- Utility-class with static methods to
find all sub-classes of a certain class (or interface) within the scope
of the current class-loader
find all sub-classes of a certain class (or interface) within the scope
of the current class-loader that can be instantiated, i.e.
- SubclassFinder() -
Constructor for class de.jstacs.utils.SubclassFinder
-
- subSampling(double) -
Method in class de.jstacs.data.DataSet
- Randomly samples elements, i.e.
- subSampling(double, double[]) -
Method in class de.jstacs.data.DataSet
-
- SubTensor - Class in de.jstacs.algorithms.graphs.tensor
- This Tensor can be used to extract or use only a part of a complete
Tensor. - SubTensor(Tensor, int, int) -
Constructor for class de.jstacs.algorithms.graphs.tensor.SubTensor
- This constructor creates a
SubTensor using the Tensor t for the nodes offset, offset+1, ..., offset+length-1.
- sum -
Variable in class de.jstacs.classifiers.differentiableSequenceScoreBased.AbstractOptimizableFunction
- The sums of the weighted data per class and additional the total weight
sum.
- sum(double[]) -
Static method in class de.jstacs.sequenceScores.statisticalModels.differentiable.directedGraphicalModels.structureLearning.measures.Measure
- Computes the sum of all elements in the array
ar.
- sum(double...) -
Static method in class de.jstacs.utils.ToolBox
- Computes the sum of the values in
array
- sum(int, int, double[]) -
Static method in class de.jstacs.utils.ToolBox
- Computes the sum of the values in
array starting at
start until end.
- sumNormalisation(double[]) -
Static method in class de.jstacs.utils.Normalisation
- The method does a sum-normalisation on
d, i.e.
- sumNormalisation(double[], double[], int) -
Static method in class de.jstacs.utils.Normalisation
- The method does a sum-normalisation on
d, i.e.
- swap() -
Method in class de.jstacs.sequenceScores.statisticalModels.trainable.mixture.AbstractMixtureTrainSM
- This method swaps the current component models with the alternative
model.
- symbol -
Variable in class de.jstacs.sequenceScores.statisticalModels.differentiable.directedGraphicalModels.BNDiffSMParameter
- The symbol (out of some
Alphabet) this parameter
is responsible for.
- SymbolExtractor - Class in de.jstacs.io
- This class enables you to extract elements (symbols) from a given
String similar to a StringTokenizer. - SymbolExtractor(String) -
Constructor for class de.jstacs.io.SymbolExtractor
- Creates a new
SymbolExtractor using delim as
delimiter.
- SymbolExtractor(String, String) -
Constructor for class de.jstacs.io.SymbolExtractor
- Creates a new
SymbolExtractor using delim as
delimiter and string as the String to be parsed.
- SymmetricTensor - Class in de.jstacs.algorithms.graphs.tensor
- This class can be used for
Tensors with a special symmetry property. - SymmetricTensor(int, byte) -
Constructor for class de.jstacs.algorithms.graphs.tensor.SymmetricTensor
- This constructor creates an empty symmetric tensor with given dimension.
- SymmetricTensor(SymmetricTensor[], double[]) -
Constructor for class de.jstacs.algorithms.graphs.tensor.SymmetricTensor
- The constructor can be used creating a new
SymmetricTensor as
weighted sum of SymmetricTensors.
- SymmetricTensor(AsymmetricTensor) -
Constructor for class de.jstacs.algorithms.graphs.tensor.SymmetricTensor
- This constructor creates and checks a filled asymmetric tensor from an
AsymmetricTensor instance.
- SymmetricTensor(double[][][], int, byte) -
Constructor for class de.jstacs.algorithms.graphs.tensor.SymmetricTensor
- This constructor creates and checks a filled asymmetric tensor with given
dimension.