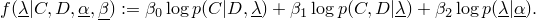BurnInTest
lastParameters
Alphabet.
0 for
arbitrary length
StartDistanceForecaster that uses the
median of a limited memory over the last values.slots memory slots
that will initially be filled with value.
AbstractStringExtractor allows to read only a limited amount of data.AbstractStringExtractor and a maximal number of Strings to be read.
Parameters from the leaves of this tree in
left-to-right order (as specified by the order of the alphabet) and
returns them as a LinkedList.
ResultSets that are part of this
ListResult
Result that contains a list or a matrix, respectively, of
ResultSets.ListResult from an array of ResultSets and
a ResultSet of annotations, which may provide additional
information on this ListResult.
Storable.
ParameterSet.
SequenceAnnotation that has a position on the sequence,
e.g for transcription start sites or intron-exon junctions.LocatedSequenceAnnotation of type type
with identifier identifier and additional annotation (that
does not fit the SequenceAnnotation definitions) given as an
array of Results result.
LocatedSequenceAnnotation of type type
with identifier identifier and additional annotation (that
does not fit the SequenceAnnotation definitions) given as a
Collection of Results result.
LocatedSequenceAnnotation of type type
with identifier identifier, additional annotation (that does
not fit the SequenceAnnotation definitions) given as an array of
Results additionalAnnotation and sub-annotations
annotations.
LocatedSequenceAnnotation of type type
with identifier identifier, additional annotation (that does
not fit the SequenceAnnotation definitions) given as an array of
Results additionalAnnotation and sub-annotations
annotations.
Storable.
SequenceAnnotation that has a position on the sequence
and a length, e.g. for donor splice sites, exons or genes.LocatedSequenceAnnotationWithLength of type
type with identifier identifier and additional
annotation (that does not fit the SequenceAnnotation definitions)
given as an array of Results result.
LocatedSequenceAnnotationWithLength of type
type with identifier identifier and additional
annotation (that does not fit the SequenceAnnotation definitions)
given as a Collection of Results result.
LocatedSequenceAnnotationWithLength of type
type with identifier identifier, additional
annotation (that does not fit the SequenceAnnotation definitions)
given as an array of Results additionalAnnotations
and sub-annotations annotations.
LocatedSequenceAnnotationWithLength of type
type with identifier identifier, additional
annotation (that does not fit the SequenceAnnotation definitions)
given as an array of Results additionalAnnotations
and sub-annotations annotations.
Storable.
double contains the sum of the logarithm of the gamma
functions used in the prior.
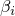 have to sum to 1.
have to sum to 1.Optimizer.
double contains the logarithm of the normalization
constant of hidden parameters of the instance.
Sequence in the given interval.
d with the values
of d given as d[i] = Math.log( val[i] ).
d between start
index startD and end index endD with the values
of d given as d[i] = Math.log( val[i] ).
d within start
index startD and end index endD with the values
of d given as d[i] = Math.log( val[i] ).
d between start
index startD and end index endD with the values
of d given as d[i] = Math.log( val[i] ).
d within start
index startD and end index endD with the values
of d given logarithmised:
d[i] = Math.log( val[i] ).
d with the values
of d given as d[i] = Math.log( val[i] ) using
offset offset.
d between start
index startD and end index endD with the values
of d given as d[i] = Math.log( val[i] ) using
offset offset.
d between start
index startD and end index endD with the values
of d given as d[i] = Math.log( val[i] ) using
offset offset.