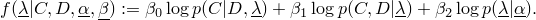Parameters and ParameterSets and the parameter representation
in Galaxy.GalaxyAdaptor from a given ParameterSet containing all parameters
that are necessary for a program is shall be included in a Galaxy installation.
Result for files that are results of some computation.GalaxyAdaptor.FileResult with name, comment, and path to the file.
GalaxyAdaptor.FileResult with name, comment, path to the file, filename and extension.
GalaxyAdaptor.FileResult from its XML-representation
ImageResult that is linked to a file that can be downloaded.ImageResult with linked GalaxyAdaptor.FileResult link
GalaxyAdaptor.LinkedImageResult from its XML-representation
Parameters that can be converted to and extracted from
Galaxy representations.GaussianEmission which can be used for maximum likelihood.
GaussianEmission with normal-gamma prior by directly defining the hyper-parameters of the prior.
GaussianEmission with normal-gamma prior by defining the expected precision and the expected standard deviation of the precision, i.e. via the
expectation and variance of the gamma part of the normal-gamma prior.
GaussianEmission from its XML representation.
Storable.
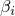 have to sum to 1.
have to sum to 1.genBeta, disBeta, and priorBeta into an array and calls the main constructor.
Storable.
GenDisMixClassifier.Storable.
GenDisMixClassifierParameterSet.
GenDisMixClassifierParameterSet.
n with entries 1/n.
number of array entries to
1/number.
n-dimensional random array.
n-dimensional random array as part of the array
d beginning at start.
number with one entry getting
the value 1-epsilon and all the others equal parts of
epsilon.
number as part of the array
d beginning at index start with one entry
getting the value 1-epsilon and all the others equal parts
of epsilon.
Storable.
GenericComplementableDiscreteAlphabet from a parameter set.
GenericComplementableDiscreteAlphabet.GenericComplementableDiscreteAlphabet.
Storable.
index.
index.
k-mers
in the data.
k-mers
in the data.
StringBuffer containing additional
information for the XML representation.
String with index index.
s1 and s2
(Alignment.Alignment(AlignmentType, Costs)).
s1 and s2
(Alignment.Alignment(AlignmentType, Costs)).
Sequences containing all elements of this
Sample.
PhyloNodes that are leafs in the subtree starting from this instance
PhyloNodes that represent the leafs of the tree
Alphabet of position pos.
AlphabetContainer of this Sample.
AlphabetContainer, used in this
Sequence.
AlphabetContainer of the
StructureLearner.
AlphabetContainer of this emission.
AlphabetContainer of the current instance.
AlphabetContainer for this ScoringFunction.
Alphabet that is used for the given position.
Alphabet of position
pos.
Collection of parameters containing informations about
this ClassifierAssessmentAssessParameterSet.
LinkedList
of Results.
Samples.
Sample.
Sequence.
ListResult.
SequenceAnnotation as
given in the constructor.
SequenceAnnotation[] for each dimension of this multidimensional sequence.
SequenceAnnotation types and the corresponding
identifier which occur in this Sample.
k nodes from the (encoded) set par to the node
child.
Sample containing the predicted binding sites.
Sample containing the predicted binding sites.
boolean which is the value of the
Parameter par.
byte which is the value of the Parameter
par.
byte array.
PhyloNodes that are children of this instance
Results of dimension
AbstractClassifier.getNumberOfClasses() that contains information about the
classifier and for each class.
GenDisMixClassifier, where the parameters
are set to those that yielded the maximum value of the objective functions among all sampled
parameter values.
GenDisMixClassifier, where the parameters
are set to the mean values over all sampled
parameter values in the stationary phase.
Storable corresponding to
the XML representation stored in this StorableResult.
index.
AbstractScoreBasedClassifier.
fgStats and
bgStats counted on sequences with a total weight of
n.
fgStats and
bgStats counted on sequences with a total weight of
nFg and nBg, respectively.
sym of the Alphabet
of position pos of this AlphabetContainer.
CollectionParameter that contains
InstanceParameterSet for each possible
class.
CollectionParameter that allows the user to choose
between different scales.
Parameter that tells something about
useful values, domains, usage of this parameter, etc.
Result.
MeasureParameters.Measure.
motifLength so that each String is contained in all
sequences of the sample respectively in the sample and the reverse
complementary sample.
code.
AlphabetContainer of Alphabets e.g. for
composite motifs/sequences.
Sequences of all
elements in the current Sample.
Sample of
Sequence.CompositeSequences.
Sequence.CompositeSequence for
sequences with a simple AlphabetContainer.
Sequences.
String representation of the context.
index.
s1(i) and
s2(j) coming from from.
index.
ConfusionMatrix.
SequenceAnnotation.
ParameterSet of the classifier.
InstanceParameterSet that has been used to
instantiate the current instance of the implementing class.
double array of dimension
ScoringFunction.getNumberOfParameters() containing the current parameter values.
SequenceAnnotation or null if no SequenceAnnotation is available.
OptimizableFunction.
Sample.PartitionMethod defining how the mutually exclusive
random-splits of user supplied data are generated.
Sample.PartitionMethod defining how the mutually exclusive
random-splits of user supplied data are generated.
Sample.PartitionMethod defining how the mutually exclusive
random-splits of user supplied data are generated.
Parameter.
Result.
true if the temporary parameter files shall
be deleted on exit of the program.
index
i.
Function.
DinucleotideProperty as a two-dimensional double array, where the rows correspond to the first nucleotide
and the columns correspond to the second nucleotide in the dinucleotide in order A, C, G, and T.
double which is the value of the
Parameter par.
fgStats and
bgStats counted on sequences with a total weight of
nFg and nBg, respectively.
fgStats and
bgStats counted on sequences with a total weight of
nFg and nBg, respectively.
String number idx that has been extracted.
Sequence, with index
i.
Sequence with index index.
ClassifierAssessmentAssessParameterSet.
Sequences, in this
Sample.
String indentifier for the type of this state.
String representation of this instance in the method Sequence.toString(String, int, int).
LocatedSequenceAnnotationWithLength, i.e.
null if no range was specified.
Constraint as exact as
possible.
null if the constraint was fulfilled by the last checked
value.
ParameterValidator.checkValue(Object) returned false.
StructureLearner.
ClassifierAssessmentAssessParameterSet.
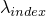 at position
at position
index: 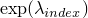 .
.
Math.exp(Parameter.getValue()), which is pre-computed.
QuadraticFunction.
AbstractHMM.fillLogStatePosteriorMatrix(double[][], int, int, Sequence, boolean) is used with code>silentZero==true
to eliminate the first row.
ParameterTree in the topological ordering of the network
structure of the enclosing BayesianNetworkScoringFunction.
float which is the value of the
Parameter par.
true if only free parameters shall be used
index.
Sequence seq beginning at position
start.
NormalizableScoringFunction.
NormalizableScoringFunction.
StringBuffer.
AbstractBurnInTest.
StringBuffer.
length.
motif used in
component.
String representation of the structure that
can be used in Graphviz to create an image.
String representation of the node options that
can be used in Graphviz to create the node for this state.
String representation of the structure that
can be used in Graphviz to create an image.
String representation of the structure that
can be used in Graphviz to create an image.
String representation of the structure that
can be used in Graphviz to create an image.
Sequence and seq.
i of the hyperparameter vector
of the underlying Dirichlet distribution.
i of the hyperparameter vector
of the underlying Erlang distribution.
index.
Parameter.
ParameterSet.
SequenceAnnotation as given in the
constructor.
NullProgressUpdater that is
immutable.
sortedScores for the index
i so that
sortedScores[i-1] < myScore <= sortedScores[i].
sortedScores beginning at
start for the index i so that
sortedScores[i-1] < myScore <= sortedScores[i].
combi.
pos - 1 to pos in sequences seq.
pos - 1 to pos in sequences seq.
String) for your
choice.
Sequences to specific Alphabets.
i of the component with
P(i|s) maximal.
- getIndexOfMaximalComponentFor(Sequence) -
Method in interface de.jstacs.motifDiscovery.MotifDiscoverer
- Returns the index of the component with the maximal score for the
sequence
sequence.
- getIndexOfMaximalComponentFor(Sequence) -
Method in class de.jstacs.scoringFunctions.IndependentProductScoringFunction
-
- getIndexOfMaximalComponentFor(Sequence) -
Method in class de.jstacs.scoringFunctions.MappingScoringFunction
-
- getIndexOfMaximalComponentFor(Sequence, int) -
Method in class de.jstacs.scoringFunctions.mix.AbstractMixtureScoringFunction
- Returns the index of the component that has the greatest impact on the
complete score for a
Sequence.
- getIndexOfMaximalComponentFor(Sequence) -
Method in class de.jstacs.scoringFunctions.mix.MixtureScoringFunction
-
- getIndexOfMaximalComponentFor(Sequence) -
Method in class de.jstacs.scoringFunctions.mix.motifSearch.HiddenMotifsMixture
-
- getIndices() -
Method in class de.jstacs.scoringFunctions.IndependentProductScoringFunction
- This method returns a deep copy of the internally used indices of the
NormalizableScoringFunction for the parts.
- getIndices(int) -
Method in class de.jstacs.scoringFunctions.mix.AbstractMixtureScoringFunction
- This array is used to compute the relative indices of a parameter index.
- getInfixSample(int, int) -
Method in class de.jstacs.data.Sample
- This method enables you to use only an infix of all elements, i.e. the
Sequences, in the current Sample.
- getInfos() -
Method in class de.jstacs.results.MeanResultSet
- Returns some information for this
MeanResultSet.
- getInitialClassParam(double) -
Method in class de.jstacs.models.hmm.models.DifferentiableHigherOrderHMM
-
- getInitialClassParam(double) -
Method in class de.jstacs.scoringFunctions.AbstractNormalizableScoringFunction
-
- getInitialClassParam(double) -
Method in interface de.jstacs.scoringFunctions.NormalizableScoringFunction
-
- getInitialClassParam(double) -
Method in interface de.jstacs.scoringFunctions.ScoringFunction
- Returns the initial class parameter for the class this
ScoringFunction is responsible for, based on the class
probability classProb.
- getInstance() -
Method in class de.jstacs.parameters.InstanceParameterSet
- Returns a new instance of the class of
InstanceParameterSet.getInstanceClass() that
was created using this ParameterSet.
- getInstanceClass() -
Method in class de.jstacs.parameters.InstanceParameterSet
- Returns the class of the instances that can be constructed using this
set.
- getInstanceComment() -
Method in class de.jstacs.algorithms.optimization.termination.AbsoluteValueCondition.AbsoluteValueConditionParameterSet
- Deprecated.
- getInstanceComment() -
Method in class de.jstacs.algorithms.optimization.termination.CombinedCondition.CombinedConditionParameterSet
-
- getInstanceComment() -
Method in class de.jstacs.algorithms.optimization.termination.IterationCondition.IterationConditionParameterSet
-
- getInstanceComment() -
Method in class de.jstacs.algorithms.optimization.termination.SmallDifferenceOfFunctionEvaluationsCondition.SmallDifferenceOfFunctionEvaluationsConditionParameterSet
-
- getInstanceComment() -
Method in class de.jstacs.algorithms.optimization.termination.SmallGradientConditon.SmallGradientConditonParameterSet
-
- getInstanceComment() -
Method in class de.jstacs.algorithms.optimization.termination.SmallStepCondition.SmallStepConditionParameterSet
-
- getInstanceComment() -
Method in class de.jstacs.algorithms.optimization.termination.TimeCondition.TimeConditionParameterSet
-
- getInstanceComment() -
Method in class de.jstacs.classifier.scoringFunctionBased.sampling.SamplingGenDisMixClassifierParameterSet
-
- getInstanceComment() -
Method in class de.jstacs.classifier.scoringFunctionBased.ScoreClassifierParameterSet
-
- getInstanceComment() -
Method in class de.jstacs.data.AlphabetContainerParameterSet.AlphabetArrayParameterSet
- Returns a descriptive comment on this
AlphabetContainerParameterSet.AlphabetArrayParameterSet.
- getInstanceComment() -
Method in class de.jstacs.data.AlphabetContainerParameterSet
-
- getInstanceComment() -
Method in class de.jstacs.data.AlphabetContainerParameterSet.SectionDefinedAlphabetParameterSet
- Returns a descriptive comment on this
AlphabetContainerParameterSet.SectionDefinedAlphabetParameterSet.
- getInstanceComment() -
Method in class de.jstacs.data.alphabets.ContinuousAlphabet.ContinuousAlphabetParameterSet
-
- getInstanceComment() -
Method in class de.jstacs.data.alphabets.DiscreteAlphabet.DiscreteAlphabetParameterSet
-
- getInstanceComment() -
Method in class de.jstacs.data.alphabets.DNAAlphabet.DNAAlphabetParameterSet
-
- getInstanceComment() -
Method in class de.jstacs.models.discrete.homogeneous.parameters.HomMMParameterSet
-
- getInstanceComment() -
Method in class de.jstacs.models.discrete.inhomogeneous.parameters.BayesianNetworkModelParameterSet
-
- getInstanceComment() -
Method in class de.jstacs.models.discrete.inhomogeneous.parameters.FSDAGMParameterSet
-
- getInstanceComment() -
Method in class de.jstacs.parameters.InstanceParameterSet
- Returns a comment (a textual description) of the class that can be
constructed using this
ParameterSet.
- getInstanceComment() -
Method in class de.jstacs.sampling.VarianceRatioBurnInTestParameterSet
-
- getInstanceComment() -
Method in class de.jstacs.scoringFunctions.directedGraphicalModels.BayesianNetworkScoringFunctionParameterSet
-
- getInstanceComment() -
Method in class de.jstacs.scoringFunctions.directedGraphicalModels.structureLearning.measures.btMeasures.BTExplainingAwayResidual.BTExplainingAwayResidualParameterSet
-
- getInstanceComment() -
Method in class de.jstacs.scoringFunctions.directedGraphicalModels.structureLearning.measures.btMeasures.BTMutualInformation.BTMutualInformationParameterSet
-
- getInstanceComment() -
Method in class de.jstacs.scoringFunctions.directedGraphicalModels.structureLearning.measures.InhomogeneousMarkov.InhomogeneousMarkovParameterSet
-
- getInstanceComment() -
Method in class de.jstacs.scoringFunctions.directedGraphicalModels.structureLearning.measures.pmmMeasures.PMMExplainingAwayResidual.PMMExplainingAwayResidualParameterSet
-
- getInstanceComment() -
Method in class de.jstacs.scoringFunctions.directedGraphicalModels.structureLearning.measures.pmmMeasures.PMMMutualInformation.PMMMutualInformationParameterSet
-
- getInstanceFromParameterSet(InstanceParameterSet) -
Static method in class de.jstacs.io.ParameterSetParser
- Returns an instance of a subclass of
InstantiableFromParameterSet
that can be instantiated by the InstanceParameterSet
pars.
- getInstanceFromParameterSet(ParameterSet, Class) -
Static method in class de.jstacs.io.ParameterSetParser
- Returns an instance of a subclass of
InstantiableFromParameterSet
that can be instantiated by the ParameterSet pars.
- getInstanceName() -
Method in class de.jstacs.algorithms.optimization.termination.AbsoluteValueCondition.AbsoluteValueConditionParameterSet
- Deprecated.
- getInstanceName() -
Method in class de.jstacs.algorithms.optimization.termination.CombinedCondition.CombinedConditionParameterSet
-
- getInstanceName() -
Method in class de.jstacs.algorithms.optimization.termination.IterationCondition.IterationConditionParameterSet
-
- getInstanceName() -
Method in class de.jstacs.algorithms.optimization.termination.SmallDifferenceOfFunctionEvaluationsCondition.SmallDifferenceOfFunctionEvaluationsConditionParameterSet
-
- getInstanceName() -
Method in class de.jstacs.algorithms.optimization.termination.SmallGradientConditon.SmallGradientConditonParameterSet
-
- getInstanceName() -
Method in class de.jstacs.algorithms.optimization.termination.SmallStepCondition.SmallStepConditionParameterSet
-
- getInstanceName() -
Method in class de.jstacs.algorithms.optimization.termination.TimeCondition.TimeConditionParameterSet
-
- getInstanceName() -
Method in class de.jstacs.classifier.AbstractClassifier
- Returns a short description of the classifier.
- getInstanceName() -
Method in class de.jstacs.classifier.MappingClassifier
-
- getInstanceName() -
Method in class de.jstacs.classifier.modelBased.ModelBasedClassifier
-
- getInstanceName() -
Method in class de.jstacs.classifier.scoringFunctionBased.gendismix.GenDisMixClassifier
-
- getInstanceName() -
Method in class de.jstacs.classifier.scoringFunctionBased.logPrior.CompositeLogPrior
-
- getInstanceName() -
Method in class de.jstacs.classifier.scoringFunctionBased.logPrior.DoesNothingLogPrior
-
- getInstanceName() -
Method in class de.jstacs.classifier.scoringFunctionBased.logPrior.LogPrior
- Returns a short instance name.
- getInstanceName() -
Method in class de.jstacs.classifier.scoringFunctionBased.logPrior.SeparateGaussianLogPrior
-
- getInstanceName() -
Method in class de.jstacs.classifier.scoringFunctionBased.logPrior.SeparateLaplaceLogPrior
-
- getInstanceName() -
Method in class de.jstacs.classifier.scoringFunctionBased.logPrior.SimpleGaussianSumLogPrior
-
- getInstanceName() -
Method in class de.jstacs.classifier.scoringFunctionBased.msp.MSPClassifier
-
- getInstanceName() -
Method in class de.jstacs.classifier.scoringFunctionBased.sampling.SamplingGenDisMixClassifierParameterSet
-
- getInstanceName() -
Method in class de.jstacs.classifier.scoringFunctionBased.sampling.SamplingScoreBasedClassifier
-
- getInstanceName() -
Method in class de.jstacs.classifier.scoringFunctionBased.ScoreClassifier
-
- getInstanceName() -
Method in class de.jstacs.classifier.scoringFunctionBased.ScoreClassifierParameterSet
-
- getInstanceName() -
Method in class de.jstacs.data.Alphabet.AlphabetParameterSet
-
- getInstanceName() -
Method in class de.jstacs.data.AlphabetContainerParameterSet.AlphabetArrayParameterSet
- Returns a descriptive name for this
AlphabetContainerParameterSet.AlphabetArrayParameterSet
.
- getInstanceName() -
Method in class de.jstacs.data.AlphabetContainerParameterSet
-
- getInstanceName() -
Method in class de.jstacs.data.AlphabetContainerParameterSet.SectionDefinedAlphabetParameterSet
- Returns a descriptive name for this
AlphabetContainerParameterSet.SectionDefinedAlphabetParameterSet.
- getInstanceName() -
Method in class de.jstacs.models.CompositeModel
-
- getInstanceName() -
Method in class de.jstacs.models.discrete.DGMParameterSet
-
- getInstanceName() -
Method in class de.jstacs.models.discrete.homogeneous.HomogeneousMM
-
- getInstanceName() -
Method in class de.jstacs.models.discrete.inhomogeneous.BayesianNetworkModel
-
- getInstanceName() -
Method in class de.jstacs.models.discrete.inhomogeneous.FSDAGModel
-
- getInstanceName() -
Method in class de.jstacs.models.discrete.inhomogeneous.shared.SharedStructureClassifier
-
- getInstanceName() -
Method in class de.jstacs.models.discrete.inhomogeneous.shared.SharedStructureMixture
-
- getInstanceName() -
Method in class de.jstacs.models.hmm.models.DifferentiableHigherOrderHMM
-
- getInstanceName() -
Method in class de.jstacs.models.hmm.models.HigherOrderHMM
-
- getInstanceName() -
Method in class de.jstacs.models.hmm.models.SamplingHigherOrderHMM
-
- getInstanceName() -
Method in class de.jstacs.models.hmm.models.SamplingPhyloHMM
-
- getInstanceName() -
Method in class de.jstacs.models.mixture.AbstractMixtureModel
-
- getInstanceName() -
Method in class de.jstacs.models.mixture.motif.HiddenMotifMixture
-
- getInstanceName() -
Method in class de.jstacs.models.mixture.motif.positionprior.GaussianLikePositionPrior
-
- getInstanceName() -
Method in class de.jstacs.models.mixture.motif.positionprior.PositionPrior
- Returns the instance name.
- getInstanceName() -
Method in class de.jstacs.models.mixture.motif.positionprior.UniformPositionPrior
-
- getInstanceName() -
Method in interface de.jstacs.models.Model
- Should return a short instance name such as iMM(0), BN(2), ...
- getInstanceName() -
Method in class de.jstacs.models.NormalizableScoringFunctionModel
-
- getInstanceName() -
Method in class de.jstacs.models.UniformModel
-
- getInstanceName() -
Method in class de.jstacs.models.VariableLengthWrapperModel
-
- getInstanceName() -
Method in class de.jstacs.parameters.InstanceParameterSet
- Returns the name of an instance of the class that can be constructed
using this
ParameterSet.
- getInstanceName() -
Method in interface de.jstacs.sampling.BurnInTest
- Returns a short description of the burn-in test.
- getInstanceName() -
Method in class de.jstacs.sampling.SimpleBurnInTest
- Deprecated.
- getInstanceName() -
Method in class de.jstacs.sampling.VarianceRatioBurnInTest
-
- getInstanceName() -
Method in class de.jstacs.sampling.VarianceRatioBurnInTestParameterSet
-
- getInstanceName() -
Method in class de.jstacs.scoringFunctions.CMMScoringFunction
-
- getInstanceName() -
Method in class de.jstacs.scoringFunctions.directedGraphicalModels.BayesianNetworkScoringFunction
-
- getInstanceName() -
Method in class de.jstacs.scoringFunctions.directedGraphicalModels.BayesianNetworkScoringFunctionParameterSet
-
- getInstanceName() -
Method in class de.jstacs.scoringFunctions.directedGraphicalModels.structureLearning.measures.btMeasures.BTExplainingAwayResidual.BTExplainingAwayResidualParameterSet
-
- getInstanceName() -
Method in class de.jstacs.scoringFunctions.directedGraphicalModels.structureLearning.measures.btMeasures.BTExplainingAwayResidual
-
- getInstanceName() -
Method in class de.jstacs.scoringFunctions.directedGraphicalModels.structureLearning.measures.btMeasures.BTMutualInformation.BTMutualInformationParameterSet
-
- getInstanceName() -
Method in class de.jstacs.scoringFunctions.directedGraphicalModels.structureLearning.measures.btMeasures.BTMutualInformation
-
- getInstanceName() -
Method in class de.jstacs.scoringFunctions.directedGraphicalModels.structureLearning.measures.InhomogeneousMarkov
-
- getInstanceName() -
Method in class de.jstacs.scoringFunctions.directedGraphicalModels.structureLearning.measures.InhomogeneousMarkov.InhomogeneousMarkovParameterSet
-
- getInstanceName() -
Method in class de.jstacs.scoringFunctions.directedGraphicalModels.structureLearning.measures.Measure
- Returns the name of the
Measure and possibly some additional
information about the current instance.
- getInstanceName() -
Method in class de.jstacs.scoringFunctions.directedGraphicalModels.structureLearning.measures.pmmMeasures.PMMExplainingAwayResidual
-
- getInstanceName() -
Method in class de.jstacs.scoringFunctions.directedGraphicalModels.structureLearning.measures.pmmMeasures.PMMExplainingAwayResidual.PMMExplainingAwayResidualParameterSet
-
- getInstanceName() -
Method in class de.jstacs.scoringFunctions.directedGraphicalModels.structureLearning.measures.pmmMeasures.PMMMutualInformation
-
- getInstanceName() -
Method in class de.jstacs.scoringFunctions.directedGraphicalModels.structureLearning.measures.pmmMeasures.PMMMutualInformation.PMMMutualInformationParameterSet
-
- getInstanceName() -
Method in class de.jstacs.scoringFunctions.homogeneous.HMM0ScoringFunction
-
- getInstanceName() -
Method in class de.jstacs.scoringFunctions.homogeneous.HMMScoringFunction
-
- getInstanceName() -
Method in class de.jstacs.scoringFunctions.homogeneous.UniformHomogeneousScoringFunction
-
- getInstanceName() -
Method in class de.jstacs.scoringFunctions.IndependentProductScoringFunction
-
- getInstanceName() -
Method in class de.jstacs.scoringFunctions.MappingScoringFunction
-
- getInstanceName() -
Method in class de.jstacs.scoringFunctions.mix.MixtureScoringFunction
-
- getInstanceName() -
Method in class de.jstacs.scoringFunctions.mix.motifSearch.HiddenMotifsMixture
-
- getInstanceName() -
Method in class de.jstacs.scoringFunctions.mix.motifSearch.MixtureDuration
-
- getInstanceName() -
Method in class de.jstacs.scoringFunctions.mix.motifSearch.SkewNormalLikeScoringFunction
-
- getInstanceName() -
Method in class de.jstacs.scoringFunctions.mix.motifSearch.UniformDurationScoringFunction
-
- getInstanceName() -
Method in class de.jstacs.scoringFunctions.mix.StrandScoringFunction
-
- getInstanceName() -
Method in class de.jstacs.scoringFunctions.MRFScoringFunction
-
- getInstanceName() -
Method in class de.jstacs.scoringFunctions.NormalizedScoringFunction
-
- getInstanceName() -
Method in interface de.jstacs.scoringFunctions.ScoringFunction
- Returns a short instance name.
- getInstanceName() -
Method in class de.jstacs.scoringFunctions.UniformScoringFunction
-
- getInstanceParameterSets() -
Method in enum de.jstacs.data.AlphabetContainer.AlphabetContainerType
- This method returns a
LinkedList of
InstanceParameterSets which can be used to create
Alphabets that can be used in a AlphabetContainer of
the given AlphabetContainer.AlphabetContainerType.
- getInstanceParameterSets(Class<T>, String) -
Static method in class de.jstacs.utils.SubclassFinder
- This method returns a list of
InstanceParameterSets that can be used to create a subclass of clazz.
- getInternalPosition(int[]) -
Method in class de.jstacs.scoringFunctions.mix.motifSearch.PositionScoringFunction
- Copies the current value of the internal iterator in the given array.
- getIntFromParameter(Parameter) -
Static method in class de.jstacs.io.ParameterSetParser
- Returns the
int which is the value of the Parameter
par.
- getK() -
Method in class de.jstacs.classifier.assessment.KFoldCVAssessParameterSet
- Returns the number of mutually exclusive random-splits of user supplied
data defined by this
KFoldCVAssessParameterSet.
- getKLDivergence(Model, Model, int) -
Static method in class de.jstacs.models.utils.ModelTester
- Returns the Kullback-Leibler-divergence
D(p_m1||p_m2).
- getKLDivergence(double[][][]) -
Method in class de.jstacs.scoringFunctions.directedGraphicalModels.ParameterTree
- Returns the KL-divergence of the distribution of this
ParameterTree and the distribution given by
ds.
- getKLDivergence(double[], double[][][][]) -
Method in class de.jstacs.scoringFunctions.directedGraphicalModels.ParameterTree
- Returns the KL-divergence of the distribution of this
ParameterTree and a number of distribution given by
ds and weighted by weight
- getKmereSequenceStatistic(int, boolean, int, Sample...) -
Static method in class de.jstacs.motifDiscovery.KMereStatistic
- This method enables the user to get a statistic over all
k-mers
in the sequences.
- getKmereSequenceStatistic(boolean, int, HashSet<Sequence>, Sample...) -
Static method in class de.jstacs.motifDiscovery.KMereStatistic
- This method enables the user to get a statistic for a set of
k-mers.
- getLabel(String[], int) -
Method in class de.jstacs.models.hmm.transitions.BasicHigherOrderTransition.AbstractTransitionElement
- This method returns a label for the state.
- getLambda(int) -
Method in class de.jstacs.models.discrete.inhomogeneous.MEMConstraint
- Returns the value of
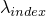 .
.
- getLastContextState(int, int) -
Method in interface de.jstacs.models.hmm.Transition
- The method returns the index of the state of the context, if there is no context -1 is returned.
- getLastContextState(int, int) -
Method in class de.jstacs.models.hmm.transitions.BasicHigherOrderTransition
-
- getLastScore() -
Method in class de.jstacs.classifier.scoringFunctionBased.ScoreClassifier
- Returns the score that was computed in the last optimization of the
parameters.
- getLegalName(String) -
Static method in class de.jstacs.utils.galaxy.GalaxyAdaptor
- Returns a legal variable name in Galaxy
- getLength() -
Method in class de.jstacs.algorithms.alignment.StringAlignment
- This method return the length of the alignment.
- getLength() -
Method in class de.jstacs.classifier.AbstractClassifier
- Returns the length of the sequences this classifier can handle or
0 for sequences of arbitrary length.
- getLength() -
Method in class de.jstacs.data.Sequence.CompositeSequence
-
- getLength() -
Method in class de.jstacs.data.Sequence
- Returns the length of the
Sequence.
- getLength() -
Method in class de.jstacs.data.Sequence.SubSequence
-
- getLength() -
Method in class de.jstacs.data.sequences.annotation.LocatedSequenceAnnotationWithLength
- Returns the length of this
LocatedSequenceAnnotationWithLength as
given in the constructor.
- getLength() -
Method in class de.jstacs.data.sequences.ArbitrarySequence
-
- getLength() -
Method in class de.jstacs.data.sequences.ByteSequence
-
- getLength() -
Method in class de.jstacs.data.sequences.IntSequence
-
- getLength() -
Method in class de.jstacs.data.sequences.MappedDiscreteSequence
-
- getLength() -
Method in class de.jstacs.data.sequences.MultiDimensionalDiscreteSequence
-
- getLength() -
Method in class de.jstacs.data.sequences.PermutedSequence
-
- getLength() -
Method in class de.jstacs.data.sequences.ShortSequence
-
- getLength() -
Method in class de.jstacs.data.sequences.SparseSequence
-
- getLength() -
Method in class de.jstacs.models.AbstractModel
-
- getLength() -
Method in class de.jstacs.models.hmm.models.HigherOrderHMM
-
- getLength() -
Method in class de.jstacs.models.mixture.motif.positionprior.GaussianLikePositionPrior
-
- getLength() -
Method in class de.jstacs.models.mixture.motif.positionprior.PositionPrior
- Returns the length that is supported by this prior.
- getLength() -
Method in class de.jstacs.models.mixture.motif.positionprior.UniformPositionPrior
-
- getLength() -
Method in interface de.jstacs.models.Model
- Returns the length of sequences this model can classify.
- getLength() -
Method in class de.jstacs.parameters.SequenceScoringParameterSet
- Returns the length of the sequences the model can work on.
- getLength() -
Method in class de.jstacs.scoringFunctions.AbstractNormalizableScoringFunction
-
- getLength() -
Method in interface de.jstacs.scoringFunctions.ScoringFunction
- Returns the length of this
ScoringFunction, i.e. the length of
the Sequences this ScoringFunction can handle.
- getLengthOfBurnIn() -
Method in class de.jstacs.sampling.AbstractBurnInTest
-
- getLengthOfBurnIn() -
Method in interface de.jstacs.sampling.BurnInTest
- Computes and returns the length of the burn-in phase using the values
from
BurnInTest.setValue(double).
- getLengthOfBurnIn() -
Method in class de.jstacs.sampling.SimpleBurnInTest
- Deprecated.
- getLengthOfModels() -
Method in class de.jstacs.models.CompositeModel
- This method returns the length of the models in the
CompositeModel.
- getLine(int) -
Method in class de.jstacs.classifier.AbstractScoreBasedClassifier.DoubleTableResult
- Return the line with a given
index from the table.
- getLineEps() -
Method in class de.jstacs.models.hmm.training.NumericalHMMTrainingParameterSet
- This method returns the threshold that should be used for stopping the line search during the optimization.
- getLink() -
Method in class de.jstacs.utils.galaxy.GalaxyAdaptor.LinkedImageResult
- Returns the linked file
- getList() -
Method in class de.jstacs.parameters.RangeParameter
- Returns a list of all parameter values as a
String or
null if no parameter values have been set.
- getLnFreq(int) -
Method in class de.jstacs.models.discrete.homogeneous.HomogeneousModel.HomCondProb
- Returns the logarithmic frequency at a given position
index.
- getLnFreq(int) -
Method in class de.jstacs.models.discrete.inhomogeneous.InhCondProb
- Returns the logarithm of the relative frequency (=probability) at
position
index in the distribution.
- getLnFreq(Sequence, int) -
Method in class de.jstacs.models.discrete.inhomogeneous.InhCondProb
- Returns the logarithm of the relative frequency (=probability) with the
position in the distribution given by the index of the specific
constraint that is fulfilled by the
Sequence s
beginning at start.
- getLogCDF(double) -
Static method in class de.jstacs.scoringFunctions.mix.motifSearch.CDFOfNormal
- This method computes the logarithm of the cumulative density function of a standard normal distribution.
- getLogGammaScoreForCurrentStatistic() -
Method in interface de.jstacs.models.hmm.states.SamplingState
- This method calculates a score for the current statistics, which is independent from the current parameters
In general the gamma-score is a product of gamma-functions parameterized with the current statistics
- getLogGammaScoreForCurrentStatistic() -
Method in class de.jstacs.models.hmm.states.SimpleSamplingState
-
- getLogGammaScoreFromStatistic() -
Method in class de.jstacs.models.hmm.states.emissions.discrete.AbstractConditionalDiscreteEmission
-
- getLogGammaScoreFromStatistic() -
Method in class de.jstacs.models.hmm.states.emissions.discrete.PhyloDiscreteEmission
-
- getLogGammaScoreFromStatistic() -
Method in interface de.jstacs.models.hmm.states.emissions.SamplingEmission
- This method calculates a score for the current statistics, which is independent from the current parameters
In general the gamma-score is a product of gamma-functions parameterized with the current statistics
- getLogGammaScoreFromStatistic() -
Method in class de.jstacs.models.hmm.states.emissions.SilentEmission
-
- getLogGammaScoreFromStatistic() -
Method in class de.jstacs.models.hmm.transitions.BasicHigherOrderTransition.AbstractTransitionElement
- This method calculates a score for the current statistics, which is independent from the current parameters
In general the gamma-score is a product of gamma-functions parameterized with the current statistics
- getLogGammaScoreFromStatistic() -
Method in class de.jstacs.models.hmm.transitions.BasicHigherOrderTransition
-
- getLogGammaScoreFromStatistic() -
Method in class de.jstacs.models.hmm.transitions.elements.ReferenceBasedTransitionElement
-
- getLogGammaScoreFromStatistic() -
Method in interface de.jstacs.models.hmm.transitions.TransitionWithSufficientStatistic
- This method calculates a score for the current statistics, which is independent from the current parameters
In general the gamma-score is a product of gamma-functions parameterized with the current statistics
- getLogGammaSum(Constraint, double) -
Static method in class de.jstacs.models.discrete.ConstraintManager
- Computes the sum of the differences between the logarithmic values of the
prior knowledge and all counts of a
Constraint c.
- getLogLikelihood(Model, Sample) -
Static method in class de.jstacs.models.utils.ModelTester
- Returns the log-likelihood of a
Sample data for a
given model m.
- getLogLikelihood(Model, Sample, double[]) -
Static method in class de.jstacs.models.utils.ModelTester
- Returns the log-likelihood of a
Sample data for a
given model m.
- getLogNormalizationConstant() -
Method in class de.jstacs.models.hmm.models.DifferentiableHigherOrderHMM
-
- getLogNormalizationConstant() -
Method in class de.jstacs.scoringFunctions.AbstractVariableLengthScoringFunction
-
- getLogNormalizationConstant(int) -
Method in class de.jstacs.scoringFunctions.CMMScoringFunction
-
- getLogNormalizationConstant() -
Method in class de.jstacs.scoringFunctions.directedGraphicalModels.BayesianNetworkScoringFunction
-
- getLogNormalizationConstant(int) -
Method in class de.jstacs.scoringFunctions.homogeneous.HMM0ScoringFunction
-
- getLogNormalizationConstant(int) -
Method in class de.jstacs.scoringFunctions.homogeneous.HMMScoringFunction
-
- getLogNormalizationConstant(int) -
Method in class de.jstacs.scoringFunctions.homogeneous.UniformHomogeneousScoringFunction
-
- getLogNormalizationConstant() -
Method in class de.jstacs.scoringFunctions.IndependentProductScoringFunction
-
- getLogNormalizationConstant() -
Method in class de.jstacs.scoringFunctions.MappingScoringFunction
-
- getLogNormalizationConstant() -
Method in class de.jstacs.scoringFunctions.mix.AbstractMixtureScoringFunction
-
- getLogNormalizationConstant() -
Method in class de.jstacs.scoringFunctions.mix.motifSearch.DurationScoringFunction
-
- getLogNormalizationConstant(int) -
Method in class de.jstacs.scoringFunctions.mix.VariableLengthMixtureScoringFunction
-
- getLogNormalizationConstant() -
Method in class de.jstacs.scoringFunctions.MRFScoringFunction
-
- getLogNormalizationConstant() -
Method in interface de.jstacs.scoringFunctions.NormalizableScoringFunction
- Returns the logarithm of the sum of the scores over all sequences of the event space.
- getLogNormalizationConstant() -
Method in class de.jstacs.scoringFunctions.NormalizedScoringFunction
-
- getLogNormalizationConstant() -
Method in class de.jstacs.scoringFunctions.UniformScoringFunction
-
- getLogNormalizationConstant(int) -
Method in interface de.jstacs.scoringFunctions.VariableLengthScoringFunction
- This method returns the logarithm of the normalization constant for a given sequence
length.
- getLogNormalizationConstantForComponent(int) -
Method in class de.jstacs.scoringFunctions.mix.AbstractMixtureScoringFunction
- Computes the logarithm of the normalization constant for the component
i.
- getLogNormalizationConstantForComponent(int) -
Method in class de.jstacs.scoringFunctions.mix.MixtureScoringFunction
-
- getLogNormalizationConstantForComponent(int) -
Method in class de.jstacs.scoringFunctions.mix.motifSearch.HiddenMotifsMixture
-
- getLogNormalizationConstantForComponent(int) -
Method in class de.jstacs.scoringFunctions.mix.StrandScoringFunction
-
- getLogNumberOfPossibleOriginalSequences() -
Method in class de.jstacs.data.sequences.MappedDiscreteSequence
- This method returns the logarithm of the number of original
Sequences that yield the same mapped Sequence.
- getLogNumberOfPossibleOriginalSequences(int, int) -
Method in class de.jstacs.data.sequences.MappedDiscreteSequence
- This method returns the logarithm of the number of original
Sequences that yield the same mapped Sequence.
- getLogNumberOfSimilarSymbols(int) -
Method in class de.jstacs.data.alphabets.DiscreteAlphabetMapping
- This method returns the logarithm of the number of old values that yield the same new value.
- getLogPartialNormalizationConstant(int) -
Method in class de.jstacs.models.hmm.models.DifferentiableHigherOrderHMM
-
- getLogPartialNormalizationConstant(int) -
Method in class de.jstacs.scoringFunctions.AbstractVariableLengthScoringFunction
-
- getLogPartialNormalizationConstant(int, int) -
Method in class de.jstacs.scoringFunctions.CMMScoringFunction
-
- getLogPartialNormalizationConstant(int) -
Method in class de.jstacs.scoringFunctions.directedGraphicalModels.BayesianNetworkScoringFunction
-
- getLogPartialNormalizationConstant(int, int) -
Method in class de.jstacs.scoringFunctions.homogeneous.HMM0ScoringFunction
-
- getLogPartialNormalizationConstant(int, int) -
Method in class de.jstacs.scoringFunctions.homogeneous.HMMScoringFunction
-
- getLogPartialNormalizationConstant(int, int) -
Method in class de.jstacs.scoringFunctions.homogeneous.UniformHomogeneousScoringFunction
-
- getLogPartialNormalizationConstant(int) -
Method in class de.jstacs.scoringFunctions.IndependentProductScoringFunction
-
- getLogPartialNormalizationConstant(int) -
Method in class de.jstacs.scoringFunctions.MappingScoringFunction
-
- getLogPartialNormalizationConstant(int) -
Method in class de.jstacs.scoringFunctions.mix.MixtureScoringFunction
-
- getLogPartialNormalizationConstant(int) -
Method in class de.jstacs.scoringFunctions.mix.motifSearch.DurationScoringFunction
-
- getLogPartialNormalizationConstant(int) -
Method in class de.jstacs.scoringFunctions.mix.motifSearch.HiddenMotifsMixture
-
- getLogPartialNormalizationConstant(int) -
Method in class de.jstacs.scoringFunctions.mix.StrandScoringFunction
-
- getLogPartialNormalizationConstant(int, int) -
Method in class de.jstacs.scoringFunctions.mix.VariableLengthMixtureScoringFunction
-
- getLogPartialNormalizationConstant(int) -
Method in class de.jstacs.scoringFunctions.MRFScoringFunction
-
- getLogPartialNormalizationConstant(int) -
Method in interface de.jstacs.scoringFunctions.NormalizableScoringFunction
- Returns the logarithm of the partial normalization constant for the parameter with index
parameterIndex.
- getLogPartialNormalizationConstant(int) -
Method in class de.jstacs.scoringFunctions.NormalizedScoringFunction
-
- getLogPartialNormalizationConstant(int) -
Method in class de.jstacs.scoringFunctions.UniformScoringFunction
-
- getLogPartialNormalizationConstant(int, int) -
Method in interface de.jstacs.scoringFunctions.VariableLengthScoringFunction
- This method returns the logarithm of the partial normalization constant for a given
parameter index and a sequence length.
- getLogPartialNormalizer() -
Method in class de.jstacs.scoringFunctions.directedGraphicalModels.Parameter
- Returns the partial derivative of the normalization constant with respect
to this parameter.
- getLogPosteriorFromStatistic() -
Method in class de.jstacs.models.hmm.models.SamplingHigherOrderHMM
- This method calculates the a posteriori probability for the current statistics
- getLogPosteriorFromStatistic() -
Method in class de.jstacs.models.hmm.states.emissions.discrete.AbstractConditionalDiscreteEmission
-
- getLogPosteriorFromStatistic() -
Method in class de.jstacs.models.hmm.states.emissions.discrete.PhyloDiscreteEmission
-
- getLogPosteriorFromStatistic() -
Method in class de.jstacs.models.hmm.states.emissions.SilentEmission
-
- getLogPosteriorFromStatistic() -
Method in class de.jstacs.models.hmm.states.SimpleSamplingState
-
- getLogPosteriorFromStatistic() -
Method in class de.jstacs.models.hmm.transitions.elements.TransitionElement
- This method computes the log posterior from the internal sufficient statistic.
- getLogPosteriorFromStatistic() -
Method in class de.jstacs.models.hmm.transitions.HigherOrderTransition
-
- getLogPosteriorFromStatistic() -
Method in interface de.jstacs.sampling.SamplingFromStatistic
- This method calculates the a-posteriori probability for the current statistics
- getLogPriorForPositions(int, int...) -
Method in class de.jstacs.models.mixture.motif.positionprior.GaussianLikePositionPrior
- Returns only the important part and leaving the logarithm of the
normalization constant out.
- getLogPriorForPositions(int, int...) -
Method in class de.jstacs.models.mixture.motif.positionprior.PositionPrior
- The logarithmic value of the prior for specified start positions of the
part motifs.
- getLogPriorForPositions(int, int...) -
Method in class de.jstacs.models.mixture.motif.positionprior.UniformPositionPrior
-
- getLogPriorTerm() -
Method in class de.jstacs.models.CompositeModel
-
- getLogPriorTerm() -
Method in class de.jstacs.models.discrete.homogeneous.HomogeneousMM
-
- getLogPriorTerm() -
Method in class de.jstacs.models.discrete.inhomogeneous.BayesianNetworkModel
-
- getLogPriorTerm() -
Method in class de.jstacs.models.discrete.inhomogeneous.DAGModel
-
- getLogPriorTerm() -
Method in class de.jstacs.models.hmm.models.HigherOrderHMM
-
- getLogPriorTerm() -
Method in class de.jstacs.models.hmm.states.emissions.continuous.GaussianEmission
-
- getLogPriorTerm() -
Method in class de.jstacs.models.hmm.states.emissions.discrete.AbstractConditionalDiscreteEmission
-
- getLogPriorTerm() -
Method in interface de.jstacs.models.hmm.states.emissions.Emission
- Returns a value that is proportional to the log of the prior.
- getLogPriorTerm() -
Method in class de.jstacs.models.hmm.states.emissions.MixtureEmission
-
- getLogPriorTerm() -
Method in class de.jstacs.models.hmm.states.emissions.SilentEmission
-
- getLogPriorTerm() -
Method in interface de.jstacs.models.hmm.Transition
- Returns a value that is proportional to the log of the prior.
- getLogPriorTerm() -
Method in class de.jstacs.models.hmm.transitions.BasicHigherOrderTransition.AbstractTransitionElement
- Returns a value that is proportional to the log of the prior.
- getLogPriorTerm() -
Method in class de.jstacs.models.hmm.transitions.BasicHigherOrderTransition
-
- getLogPriorTerm() -
Method in class de.jstacs.models.hmm.transitions.elements.ReferenceBasedTransitionElement
-
- getLogPriorTerm() -
Method in class de.jstacs.models.mixture.AbstractMixtureModel
-
- getLogPriorTerm() -
Method in interface de.jstacs.models.Model
- Returns a value that is proportional to the log of the prior.
- getLogPriorTerm() -
Method in class de.jstacs.models.NormalizableScoringFunctionModel
-
- getLogPriorTerm() -
Method in class de.jstacs.models.UniformModel
-
- getLogPriorTerm() -
Method in class de.jstacs.models.VariableLengthWrapperModel
-
- getLogPriorTerm() -
Method in class de.jstacs.scoringFunctions.CMMScoringFunction
-
- getLogPriorTerm() -
Method in class de.jstacs.scoringFunctions.directedGraphicalModels.BayesianNetworkScoringFunction
-
- getLogPriorTerm() -
Method in class de.jstacs.scoringFunctions.directedGraphicalModels.MutableMarkovModelScoringFunction
-
- getLogPriorTerm() -
Method in class de.jstacs.scoringFunctions.homogeneous.HMM0ScoringFunction
-
- getLogPriorTerm() -
Method in class de.jstacs.scoringFunctions.homogeneous.HMMScoringFunction
-
- getLogPriorTerm() -
Method in class de.jstacs.scoringFunctions.homogeneous.UniformHomogeneousScoringFunction
-
- getLogPriorTerm() -
Method in class de.jstacs.scoringFunctions.IndependentProductScoringFunction
-
- getLogPriorTerm() -
Method in class de.jstacs.scoringFunctions.MappingScoringFunction
-
- getLogPriorTerm() -
Method in class de.jstacs.scoringFunctions.mix.AbstractMixtureScoringFunction
-
- getLogPriorTerm() -
Method in class de.jstacs.scoringFunctions.mix.motifSearch.MixtureDuration
-
- getLogPriorTerm() -
Method in class de.jstacs.scoringFunctions.mix.motifSearch.SkewNormalLikeScoringFunction
-
- getLogPriorTerm() -
Method in class de.jstacs.scoringFunctions.mix.motifSearch.UniformDurationScoringFunction
-
- getLogPriorTerm() -
Method in class de.jstacs.scoringFunctions.MRFScoringFunction
-
- getLogPriorTerm() -
Method in interface de.jstacs.scoringFunctions.NormalizableScoringFunction
- This method computes a value that is proportional to
NormalizableScoringFunction.getEss() * NormalizableScoringFunction.getLogNormalizationConstant() + Math.log( prior )
where prior is the prior for the parameters of this model.
- getLogPriorTerm() -
Method in class de.jstacs.scoringFunctions.NormalizedScoringFunction
-
- getLogPriorTerm() -
Method in class de.jstacs.scoringFunctions.UniformScoringFunction
-
- getLogPriorTermForComponentProbs() -
Method in class de.jstacs.models.mixture.AbstractMixtureModel
- This method computes the part of the prior that comes from the component
probabilities.
- getLogProbAndPartialDerivationFor(boolean, int, int, IntList, DoubleList, Sequence) -
Method in class de.jstacs.models.hmm.states.emissions.continuous.GaussianEmission
-
- getLogProbAndPartialDerivationFor(boolean, int, int, IntList, DoubleList, Sequence) -
Method in interface de.jstacs.models.hmm.states.emissions.DifferentiableEmission
- Returns the logarithmic score for a
Sequence beginning at
position start in the Sequence and fills lists with
the indices and the partial derivations.
- getLogProbAndPartialDerivationFor(boolean, int, int, IntList, DoubleList, Sequence) -
Method in class de.jstacs.models.hmm.states.emissions.discrete.AbstractConditionalDiscreteEmission
-
- getLogProbAndPartialDerivationFor(boolean, int, int, IntList, DoubleList, Sequence) -
Method in class de.jstacs.models.hmm.states.emissions.discrete.PhyloDiscreteEmission
-
- getLogProbAndPartialDerivationFor(boolean, int, int, IntList, DoubleList, Sequence) -
Method in class de.jstacs.models.hmm.states.emissions.SilentEmission
-
- getLogProbFor(Sequence, int, int) -
Method in class de.jstacs.models.AbstractModel
-
- getLogProbFor(Sequence, int) -
Method in class de.jstacs.models.AbstractModel
-
- getLogProbFor(Sequence) -
Method in class de.jstacs.models.AbstractModel
-
- getLogProbFor(Sample) -
Method in class de.jstacs.models.AbstractModel
-
- getLogProbFor(Sample, double[]) -
Method in class de.jstacs.models.AbstractModel
-
- getLogProbFor(Sequence, int, int) -
Method in class de.jstacs.models.CompositeModel
-
- getLogProbFor(Sequence, int, int) -
Method in class de.jstacs.models.discrete.homogeneous.HomogeneousModel
-
- getLogProbFor(Sequence, int, int) -
Method in class de.jstacs.models.discrete.inhomogeneous.DAGModel
-
- getLogProbFor(Sequence) -
Method in class de.jstacs.models.hmm.AbstractHMM
-
- getLogProbFor(Sequence, int) -
Method in class de.jstacs.models.hmm.AbstractHMM
-
- getLogProbFor(Sequence, int, int) -
Method in class de.jstacs.models.hmm.AbstractHMM
-
- getLogProbFor(Sample) -
Method in class de.jstacs.models.hmm.models.HigherOrderHMM
-
- getLogProbFor(Sample, double[]) -
Method in class de.jstacs.models.hmm.models.HigherOrderHMM
-
- getLogProbFor(boolean, int, int, Sequence) -
Method in class de.jstacs.models.hmm.states.emissions.continuous.GaussianEmission
-
- getLogProbFor(boolean, int, int, Sequence) -
Method in class de.jstacs.models.hmm.states.emissions.discrete.AbstractConditionalDiscreteEmission
-
- getLogProbFor(boolean, int, int, Sequence) -
Method in class de.jstacs.models.hmm.states.emissions.discrete.PhyloDiscreteEmission
-
- getLogProbFor(boolean, int, int, Sequence) -
Method in interface de.jstacs.models.hmm.states.emissions.Emission
- This method computes the logarithm of the likelihood.
- getLogProbFor(boolean, int, int, Sequence) -
Method in class de.jstacs.models.hmm.states.emissions.MixtureEmission
-
- getLogProbFor(boolean, int, int, Sequence) -
Method in class de.jstacs.models.hmm.states.emissions.SilentEmission
-
- getLogProbFor(int, Sequence) -
Method in class de.jstacs.models.mixture.AbstractMixtureModel
- Returns the logarithmic probability for the sequence and the given
component.
- getLogProbFor(Sequence, int, int) -
Method in class de.jstacs.models.mixture.AbstractMixtureModel
-
- getLogProbFor(Sample) -
Method in class de.jstacs.models.mixture.AbstractMixtureModel
-
- getLogProbFor(Sequence, int, int) -
Method in interface de.jstacs.models.Model
- Returns the logarithm of the probability of (a part of) the given
sequence given the model.
- getLogProbFor(Sequence, int) -
Method in interface de.jstacs.models.Model
- Returns the logarithm of the probability of (a part of) the given
sequence given the model.
- getLogProbFor(Sequence) -
Method in interface de.jstacs.models.Model
- Returns the logarithm of the probability of the given sequence given the
model.
- getLogProbFor(Sample) -
Method in interface de.jstacs.models.Model
- This method computes the logarithm of the probabilities of all sequences
in the given sample.
- getLogProbFor(Sample, double[]) -
Method in interface de.jstacs.models.Model
- This method computes and stores the logarithm of the probabilities for
any sequence in the sample in the given
double-array.
- getLogProbFor(Sequence, int, int) -
Method in class de.jstacs.models.NormalizableScoringFunctionModel
-
- getLogProbForPath(IntList, int, Sequence) -
Method in class de.jstacs.models.hmm.AbstractHMM
-
- getLogProbForPath(IntList, int, Sequence) -
Method in class de.jstacs.models.hmm.models.HigherOrderHMM
-
- getLogProbForPath(IntList, int, Sequence) -
Method in class de.jstacs.models.hmm.models.SamplingHigherOrderHMM
-
- getLogProbUsingCurrentParameterSetFor(int, Sequence, int, int) -
Method in class de.jstacs.models.mixture.AbstractMixtureModel
- Returns the logarithmic probability for the sequence and the given
component using the current parameter set.
- getLogProbUsingCurrentParameterSetFor(int, Sequence, int, int) -
Method in class de.jstacs.models.mixture.MixtureModel
-
- getLogProbUsingCurrentParameterSetFor(int, Sequence, int, int) -
Method in class de.jstacs.models.mixture.motif.SingleHiddenMotifMixture
-
- getLogProbUsingCurrentParameterSetFor(int, Sequence, int, int) -
Method in class de.jstacs.models.mixture.StrandModel
-
- getLogProposalPosteriorFromStatistic() -
Method in class de.jstacs.models.hmm.states.emissions.discrete.PhyloDiscreteEmission
- Returns the log posterior of the proposal distribution for the current statistic
- getLogScore(Sequence) -
Method in class de.jstacs.models.hmm.models.DifferentiableHigherOrderHMM
-
- getLogScore(Sequence, int) -
Method in class de.jstacs.models.hmm.models.DifferentiableHigherOrderHMM
-
- getLogScore(Sequence) -
Method in class de.jstacs.scoringFunctions.AbstractNormalizableScoringFunction
- Returns the logarithmic score for a given
Sequence.
- getLogScore(Sequence, int) -
Method in class de.jstacs.scoringFunctions.AbstractVariableLengthScoringFunction
-
- getLogScore(Sequence, int, int) -
Method in class de.jstacs.scoringFunctions.CMMScoringFunction
-
- getLogScore(Sequence, int) -
Method in class de.jstacs.scoringFunctions.directedGraphicalModels.BayesianNetworkScoringFunction
-
- getLogScore(Sequence, int, int) -
Method in class de.jstacs.scoringFunctions.homogeneous.HMM0ScoringFunction
-
- getLogScore(Sequence, int, int) -
Method in class de.jstacs.scoringFunctions.homogeneous.HMMScoringFunction
-
- getLogScore(Sequence, int, int) -
Method in class de.jstacs.scoringFunctions.homogeneous.UniformHomogeneousScoringFunction
-
- getLogScore(Sequence, int) -
Method in class de.jstacs.scoringFunctions.IndependentProductScoringFunction
-
- getLogScore(Sequence, int) -
Method in class de.jstacs.scoringFunctions.MappingScoringFunction
-
- getLogScore(Sequence, int) -
Method in class de.jstacs.scoringFunctions.mix.AbstractMixtureScoringFunction
-
- getLogScore(Sequence, int) -
Method in class de.jstacs.scoringFunctions.mix.motifSearch.HiddenMotifsMixture
-
- getLogScore(int...) -
Method in class de.jstacs.scoringFunctions.mix.motifSearch.MixtureDuration
-
- getLogScore(int...) -
Method in class de.jstacs.scoringFunctions.mix.motifSearch.PositionScoringFunction
- This method enables the user to get the log-score without using a sequence object.
- getLogScore(Sequence, int) -
Method in class de.jstacs.scoringFunctions.mix.motifSearch.PositionScoringFunction
-
- getLogScore(int...) -
Method in class de.jstacs.scoringFunctions.mix.motifSearch.SkewNormalLikeScoringFunction
-
- getLogScore(int...) -
Method in class de.jstacs.scoringFunctions.mix.motifSearch.UniformDurationScoringFunction
-
- getLogScore(Sequence, int, int) -
Method in class de.jstacs.scoringFunctions.mix.VariableLengthMixtureScoringFunction
-
- getLogScore(Sequence, int) -
Method in class de.jstacs.scoringFunctions.MRFScoringFunction
-
- getLogScore(Sequence, int) -
Method in class de.jstacs.scoringFunctions.NormalizedScoringFunction
-
- getLogScore(Sequence) -
Method in interface de.jstacs.scoringFunctions.ScoringFunction
- Returns the logarithmic score for the
Sequence seq.
- getLogScore(Sequence, int) -
Method in interface de.jstacs.scoringFunctions.ScoringFunction
- Returns the logarithmic score for the
Sequence seq
beginning at position start in the Sequence.
- getLogScore(Sequence, int) -
Method in class de.jstacs.scoringFunctions.UniformScoringFunction
-
- getLogScore(Sequence, int, int) -
Method in interface de.jstacs.scoringFunctions.VariableLengthScoringFunction
- This method computes the logarithm of the score for a given subsequence.
- getLogScoreAndPartialDerivation(Sequence, IntList, DoubleList) -
Method in class de.jstacs.models.hmm.models.DifferentiableHigherOrderHMM
-
- getLogScoreAndPartialDerivation(Sequence, int, IntList, DoubleList) -
Method in class de.jstacs.models.hmm.models.DifferentiableHigherOrderHMM
-
- getLogScoreAndPartialDerivation(int, int, IntList, DoubleList, Sequence) -
Method in interface de.jstacs.models.hmm.states.DifferentiableState
- This method allows to compute the logarithm of the score and the gradient for the given subsequences.
- getLogScoreAndPartialDerivation(int, int, IntList, DoubleList, Sequence) -
Method in class de.jstacs.models.hmm.states.SimpleDifferentiableState
-
- getLogScoreAndPartialDerivation(int, int, int, IntList, DoubleList, Sequence, int) -
Method in interface de.jstacs.models.hmm.transitions.DifferentiableTransition
- This method allows to compute the logarithm of the score and the gradient for a specific transition.
- getLogScoreAndPartialDerivation(int, IntList, DoubleList, Sequence, int) -
Method in class de.jstacs.models.hmm.transitions.elements.TransitionElement
- Returns the logarithmic score and fills lists with
the indices and the partial derivations.
- getLogScoreAndPartialDerivation(int, int, int, IntList, DoubleList, Sequence, int) -
Method in class de.jstacs.models.hmm.transitions.HigherOrderTransition
-
- getLogScoreAndPartialDerivation(Sequence, IntList, DoubleList) -
Method in class de.jstacs.scoringFunctions.AbstractNormalizableScoringFunction
-
- getLogScoreAndPartialDerivation(Sequence, int, IntList, DoubleList) -
Method in class de.jstacs.scoringFunctions.AbstractVariableLengthScoringFunction
-
- getLogScoreAndPartialDerivation(Sequence, int, int, IntList, DoubleList) -
Method in class de.jstacs.scoringFunctions.CMMScoringFunction
-
- getLogScoreAndPartialDerivation(Sequence, int, IntList, DoubleList) -
Method in class de.jstacs.scoringFunctions.directedGraphicalModels.BayesianNetworkScoringFunction
-
- getLogScoreAndPartialDerivation(Sequence, int, int, IntList, DoubleList) -
Method in class de.jstacs.scoringFunctions.homogeneous.HMM0ScoringFunction
-
- getLogScoreAndPartialDerivation(Sequence, int, int, IntList, DoubleList) -
Method in class de.jstacs.scoringFunctions.homogeneous.HMMScoringFunction
-
- getLogScoreAndPartialDerivation(Sequence, int, int, IntList, DoubleList) -
Method in class de.jstacs.scoringFunctions.homogeneous.UniformHomogeneousScoringFunction
-
- getLogScoreAndPartialDerivation(Sequence, int, IntList, DoubleList) -
Method in class de.jstacs.scoringFunctions.IndependentProductScoringFunction
-
- getLogScoreAndPartialDerivation(Sequence, int, IntList, DoubleList) -
Method in class de.jstacs.scoringFunctions.MappingScoringFunction
-
- getLogScoreAndPartialDerivation(Sequence, int, IntList, DoubleList) -
Method in class de.jstacs.scoringFunctions.mix.MixtureScoringFunction
-
- getLogScoreAndPartialDerivation(Sequence, int, IntList, DoubleList) -
Method in class de.jstacs.scoringFunctions.mix.motifSearch.HiddenMotifsMixture
-
- getLogScoreAndPartialDerivation(IntList, DoubleList, int...) -
Method in class de.jstacs.scoringFunctions.mix.motifSearch.MixtureDuration
-
- getLogScoreAndPartialDerivation(IntList, DoubleList, int...) -
Method in class de.jstacs.scoringFunctions.mix.motifSearch.PositionScoringFunction
- This method enables the user to get the log-score and the partial derivations without using a sequence object.
- getLogScoreAndPartialDerivation(Sequence, int, IntList, DoubleList) -
Method in class de.jstacs.scoringFunctions.mix.motifSearch.PositionScoringFunction
-
- getLogScoreAndPartialDerivation(IntList, DoubleList, int...) -
Method in class de.jstacs.scoringFunctions.mix.motifSearch.SkewNormalLikeScoringFunction
-
- getLogScoreAndPartialDerivation(IntList, DoubleList, int...) -
Method in class de.jstacs.scoringFunctions.mix.motifSearch.UniformDurationScoringFunction
-
- getLogScoreAndPartialDerivation(Sequence, int, IntList, DoubleList) -
Method in class de.jstacs.scoringFunctions.mix.StrandScoringFunction
-
- getLogScoreAndPartialDerivation(Sequence, int, int, IntList, DoubleList) -
Method in class de.jstacs.scoringFunctions.mix.VariableLengthMixtureScoringFunction
-
- getLogScoreAndPartialDerivation(Sequence, int, IntList, DoubleList) -
Method in class de.jstacs.scoringFunctions.MRFScoringFunction
-
- getLogScoreAndPartialDerivation(Sequence, int, IntList, DoubleList) -
Method in class de.jstacs.scoringFunctions.NormalizedScoringFunction
-
- getLogScoreAndPartialDerivation(Sequence, IntList, DoubleList) -
Method in interface de.jstacs.scoringFunctions.ScoringFunction
- Returns the logarithmic score for a
Sequence seq and
fills lists with the indices and the partial derivations.
- getLogScoreAndPartialDerivation(Sequence, int, IntList, DoubleList) -
Method in interface de.jstacs.scoringFunctions.ScoringFunction
- Returns the logarithmic score for a
Sequence beginning at
position start in the Sequence and fills lists with
the indices and the partial derivations.
- getLogScoreAndPartialDerivation(Sequence, int, IntList, DoubleList) -
Method in class de.jstacs.scoringFunctions.UniformScoringFunction
-
- getLogScoreAndPartialDerivation(Sequence, int, int, IntList, DoubleList) -
Method in interface de.jstacs.scoringFunctions.VariableLengthScoringFunction
- This method computes the logarithm of the score and the partial
derivations for a given subsequence.
- getLogScoreAndPartialDerivationForInternal(IntList, DoubleList) -
Method in class de.jstacs.scoringFunctions.mix.motifSearch.PositionScoringFunction
- This method enables the user to get the log-score and the partial derivations without using a sequence object by using the internal iterator.
- getLogScoreFor(int, int, Sequence) -
Method in interface de.jstacs.models.hmm.State
- This method returns the logarithm of the score for a given sequence with given start and end position.
- getLogScoreFor(int, int, Sequence) -
Method in class de.jstacs.models.hmm.states.SimpleState
-
- getLogScoreFor(int, int, int, Sequence, int) -
Method in interface de.jstacs.models.hmm.Transition
- This method returns the logarithm of the score for the transition.
- getLogScoreFor(int, Sequence, int) -
Method in class de.jstacs.models.hmm.transitions.BasicHigherOrderTransition.AbstractTransitionElement
- This method returns the score for the transition from the current context to the state with index
index.
- getLogScoreFor(int, int, int, Sequence, int) -
Method in class de.jstacs.models.hmm.transitions.BasicHigherOrderTransition
-
- getLogScoreFor(int, Sequence, int) -
Method in class de.jstacs.models.hmm.transitions.elements.DistanceBasedScaledTransitionElement
-
- getLogScoreFor(int, Sequence, int) -
Method in class de.jstacs.models.hmm.transitions.elements.ScaledTransitionElement
-
- getLogScoreForInternal() -
Method in class de.jstacs.scoringFunctions.mix.motifSearch.PositionScoringFunction
- This method enables the user to get the log-score without using a sequence object by using the internal iterator.
- getLogStatePosteriorMatrixFor(int, int, Sequence) -
Method in class de.jstacs.models.hmm.AbstractHMM
- This method returns the log state posterior of all states for a sequence.
- getLogStatePosteriorMatrixFor(Sample) -
Method in class de.jstacs.models.hmm.AbstractHMM
- This method returns the log state posteriors for all sequences of the sample
data.
- getLogStatePosteriorMatrixFor(int, int, Sequence) -
Method in class de.jstacs.models.hmm.models.SamplingHigherOrderHMM
-
- getLogSum(double...) -
Static method in class de.jstacs.utils.Normalisation
- Returns the logarithm of the sum of values given as
lnVal[i] = Math.log( val[i] ).
- getLogSum(int, int, double...) -
Static method in class de.jstacs.utils.Normalisation
- Returns the logarithm of the sum of values given as
lnVal[i] = Math.log( val[i] ) between a start and end index.
- getLogT() -
Method in class de.jstacs.scoringFunctions.directedGraphicalModels.Parameter
- Returns the part of the normalization constant of parameters before this
parameter in the structure of the network.
- getLogZ() -
Method in class de.jstacs.scoringFunctions.directedGraphicalModels.Parameter
- Returns the part of the normalization constant of parameters after this
parameter in the structure of the network.
- getLongFromParameter(Parameter) -
Static method in class de.jstacs.io.ParameterSetParser
- Returns the
long which is the value of the Parameter
par.
- getLowerBound() -
Method in class de.jstacs.parameters.validation.NumberValidator
- Returns the lower bound of the
NumberValidator.
- getMarginalDistribution(Model, int[]) -
Static method in class de.jstacs.models.utils.ModelTester
- This method computes the marginal distribution for any discrete model
m and all sequences that fulfill the constraint
, if possible.
- getMarginalOrder() -
Method in class de.jstacs.models.discrete.Constraint
- Returns the marginal order, i.e. the number of used random variables.
- getMatrix() -
Method in class de.jstacs.classifier.ConfusionMatrix
- This method returns the confusion matrix as a two dimensional
int-array.
- getMax() -
Method in class de.jstacs.data.alphabets.ContinuousAlphabet
- Returns the maximal value of this alphabet.
- getMax(double[][]) -
Static method in class de.jstacs.models.discrete.inhomogeneous.TwoPointEvaluater
- This method can be used to determine the maximal value of the matrix of
mutual informations.
- getMax() -
Method in class de.jstacs.scoringFunctions.mix.motifSearch.DurationScoringFunction
- Returns the maximal value that can be scored.
- getMaximalAlphabetLength() -
Method in class de.jstacs.data.AlphabetContainer
- Returns the maximal
Alphabet length of this
AlphabetContainer.
- getMaximalEdgeFor(byte, int, int...) -
Method in class de.jstacs.algorithms.graphs.tensor.AsymmetricTensor
-
- getMaximalEdgeFor(byte, int, int...) -
Method in class de.jstacs.algorithms.graphs.tensor.SubTensor
-
- getMaximalEdgeFor(byte, int, int...) -
Method in class de.jstacs.algorithms.graphs.tensor.SymmetricTensor
-
- getMaximalEdgeFor(byte, int, int...) -
Method in class de.jstacs.algorithms.graphs.tensor.Tensor
- Returns the edge
permute(parents[0],...
- getMaximalElementLength() -
Method in class de.jstacs.data.Sample
- Returns the maximal length of an element, i.e. a
Sequence, in
this Sample.
- getMaximalInDegree() -
Method in interface de.jstacs.models.hmm.Transition
- This method returns the maximal out degree of any context used in this transition instance.
- getMaximalInDegree() -
Method in class de.jstacs.models.hmm.transitions.BasicHigherOrderTransition
-
- getMaximalMarkovOrder() -
Method in class de.jstacs.models.AbstractModel
-
- getMaximalMarkovOrder() -
Method in class de.jstacs.models.CompositeModel
-
- getMaximalMarkovOrder() -
Method in class de.jstacs.models.discrete.homogeneous.HomogeneousModel
-
- getMaximalMarkovOrder() -
Method in class de.jstacs.models.discrete.inhomogeneous.BayesianNetworkModel
-
- getMaximalMarkovOrder() -
Method in class de.jstacs.models.discrete.inhomogeneous.FSDAGModel
-
- getMaximalMarkovOrder() -
Method in class de.jstacs.models.hmm.models.HigherOrderHMM
-
- getMaximalMarkovOrder() -
Method in interface de.jstacs.models.hmm.Transition
- This method returns the maximal used Markov order.
- getMaximalMarkovOrder() -
Method in class de.jstacs.models.hmm.transitions.BasicHigherOrderTransition
-
- getMaximalMarkovOrder() -
Method in interface de.jstacs.models.Model
- This method returns the maximal used Markov order, if possible.
- getMaximalMarkovOrder() -
Method in class de.jstacs.models.UniformModel
-
- getMaximalMarkovOrder() -
Method in class de.jstacs.scoringFunctions.homogeneous.HMM0ScoringFunction
-
- getMaximalMarkovOrder() -
Method in class de.jstacs.scoringFunctions.homogeneous.HMMScoringFunction
-
- getMaximalMarkovOrder() -
Method in class de.jstacs.scoringFunctions.homogeneous.HomogeneousScoringFunction
- Returns the maximal used markov oder.
- getMaximalMarkovOrder() -
Method in class de.jstacs.scoringFunctions.homogeneous.UniformHomogeneousScoringFunction
-
- getMaximalNumberOfChildren() -
Method in interface de.jstacs.models.hmm.Transition
- This method returns the maximal number of children for any context used in this transition instance.
- getMaximalNumberOfChildren() -
Method in class de.jstacs.models.hmm.transitions.BasicHigherOrderTransition
-
- getMaximalSymbolLength() -
Method in class de.jstacs.data.alphabets.DiscreteAlphabet
- Returns the length of the longest "symbol" in the alphabet.
- getMaxIndex(double[]) -
Static method in class de.jstacs.utils.ToolBox
- Returns the index with maximal value in a
double array.
- getMaxIndex(int, int, double[]) -
Static method in class de.jstacs.utils.ToolBox
- Returns the index with maximal value in a
double array.
- getMaxOfCC(double[], double[]) -
Static method in class de.jstacs.classifier.ScoreBasedPerformanceMeasureDefinitions
- This method computes the maximal correlation coefficient (CC_max).
- getMaxOfDeviation(Model, Model, int) -
Static method in class de.jstacs.models.utils.ModelTester
- This method computes the maximum deviation between the probabilities for
all sequences of
length for discrete models m1
and m2.
- getMeanParameters(boolean, int) -
Method in class de.jstacs.classifier.scoringFunctionBased.sampling.SamplingScoreBasedClassifier
- Returns the mean parameters over all samplings of all stationary phases.
- getMeasure() -
Method in class de.jstacs.classifier.ScoreBasedPerformanceMeasureDefinitions.ThresholdMeasurePair
- This method returns the value of the measure.
- getMeasure() -
Method in class de.jstacs.scoringFunctions.directedGraphicalModels.BayesianNetworkScoringFunctionParameterSet
- Returns the structure
Measure defined by this set of parameters.
- getMeasuresForEvaluate() -
Static method in class de.jstacs.classifier.AbstractClassifier
- Returns an object of the parameters for the method
AbstractClassifier.evaluate(MeasureParameters, boolean, Sample...).
- getMeasuresForEvaluateAll() -
Static method in class de.jstacs.classifier.AbstractClassifier
- Returns an object of the parameters for the method
AbstractClassifier.evaluateAll(MeasureParameters, boolean, Sample...).
- getMI(double[][][][][][], double) -
Static method in class de.jstacs.scoringFunctions.directedGraphicalModels.structureLearning.measures.Measure
- Computes the mutual information from
counts counted on
sequences with a total weight of n.
- getMI(double[][][][], double) -
Static method in class de.jstacs.scoringFunctions.directedGraphicalModels.structureLearning.measures.Measure
- Computes the mutual information from
counts counted on
sequences with a total weight of n.
- getMIInBits(Sample, double[]) -
Static method in class de.jstacs.models.discrete.inhomogeneous.TwoPointEvaluater
- This method computes the pairwise mutual information (in bits) between
the sequence positions.
- getMin() -
Method in class de.jstacs.data.Alphabet
- Returns the minimal value of the
Alphabet.
- getMin(int) -
Method in class de.jstacs.data.AlphabetContainer
- Returns the minimal value of the underlying
Alphabet of position
pos.
- getMin() -
Method in class de.jstacs.data.alphabets.ContinuousAlphabet
-
- getMin() -
Method in class de.jstacs.data.alphabets.DiscreteAlphabet
-
- getMin() -
Method in class de.jstacs.scoringFunctions.mix.motifSearch.DurationScoringFunction
- Returns the minimal value that can be scored.
- getMinimalAlphabetLength() -
Method in class de.jstacs.data.AlphabetContainer
- Returns the minimal
Alphabet length of this
AlphabetContainer.
- getMinimalElementLength() -
Method in class de.jstacs.data.Sample
- Returns the minimal length of an element, i.e. a
Sequence, in
this Sample.
- getMinimalHyperparameter() -
Method in class de.jstacs.models.hmm.transitions.elements.TransitionElement
- This method returns the minimal hyper parameters of this
TransitionElement.
- getMinimalSequenceLength() -
Method in class de.jstacs.models.mixture.motif.HiddenMotifMixture
- Returns the minimal length a sequence respectively a sample has to have.
- getMinimalSequenceLength() -
Method in class de.jstacs.models.mixture.motif.SingleHiddenMotifMixture
-
- getMisclassificationRate() -
Method in class de.jstacs.classifier.ConfusionMatrix
- This method returns the misclassification rate.
- getModel(int) -
Method in class de.jstacs.classifier.modelBased.ModelBasedClassifier
- Returns a clone of the
Model for a specified class.
- getModel(int) -
Method in class de.jstacs.models.mixture.AbstractMixtureModel
- Returns a deep copy of the
i-th model.
- getModelInstanceName() -
Method in class de.jstacs.models.discrete.inhomogeneous.parameters.BayesianNetworkModelParameterSet
- This method returns a short description of the model.
- getModelInstanceName(StructureLearner.ModelType, byte, StructureLearner.LearningType, double) -
Static method in class de.jstacs.models.discrete.inhomogeneous.parameters.IDGMParameterSet
- This method returns a short textual representation of the model instance.
- getModels() -
Method in class de.jstacs.models.CompositeModel
- Returns the a deep copy of the models.
- getModels() -
Method in class de.jstacs.models.mixture.AbstractMixtureModel
- Returns a deep copy of the models.
- getMostProbableSequence(Model, int) -
Static method in class de.jstacs.models.utils.ModelTester
- Returns one most probable sequence for the discrete model
m.
- getMotifDiscoverer() -
Method in class de.jstacs.motifDiscovery.SignificantMotifOccurrencesFinder
- This method returns a clone of the internally used
MotifDiscoverer.
- getMotifLength(int) -
Method in class de.jstacs.models.mixture.motif.SingleHiddenMotifMixture
-
- getMotifLength(int) -
Method in interface de.jstacs.motifDiscovery.MotifDiscoverer
- This method returns the length of the motif with index
motif
.
- getMotifLength(int) -
Method in class de.jstacs.scoringFunctions.IndependentProductScoringFunction
-
- getMotifLength(int) -
Method in class de.jstacs.scoringFunctions.MappingScoringFunction
-
- getMotifLength(int) -
Method in class de.jstacs.scoringFunctions.mix.MixtureScoringFunction
-
- getMotifLength(int) -
Method in class de.jstacs.scoringFunctions.mix.motifSearch.HiddenMotifsMixture
-
- getMRG() -
Method in class de.jstacs.models.mixture.AbstractMixtureModel
- This method creates the multivariate random generator that will be used
during initialization.
- getMRGParams() -
Method in class de.jstacs.models.mixture.AbstractMixtureModel
- This method creates the parameters used in a multivariate random
generator while initialization.
- getName() -
Method in class de.jstacs.models.phylo.PhyloNode
- This method returns the name of the current instance
- getName() -
Method in class de.jstacs.models.phylo.PhyloTree
- This method returns the name of the PhyloTree
- getName() -
Method in class de.jstacs.parameters.CollectionParameter
-
- getName() -
Method in class de.jstacs.parameters.FileParameter
-
- getName() -
Method in class de.jstacs.parameters.Parameter
- Returns the name of the
Parameter.
- getName() -
Method in class de.jstacs.parameters.ParameterSetContainer
-
- getName() -
Method in class de.jstacs.parameters.RangeParameter
-
- getName() -
Method in class de.jstacs.parameters.SimpleParameter
-
- getName() -
Method in class de.jstacs.results.Result
- Returns the name of the
Result.
- getNameOfAlgorithm() -
Method in class de.jstacs.models.mixture.AbstractMixtureModel
- Returns the name of the used algorithm.
- getNameOfAssessment() -
Method in class de.jstacs.classifier.assessment.ClassifierAssessment
- Returns the name of this class.
- getNameString() -
Method in enum de.jstacs.classifier.MeasureParameters.Measure
- Returns the name of the
MeasureParameters.Measure.
- getNeededReference() -
Method in class de.jstacs.parameters.Parameter
- Returns a reference to a
ParameterSet whose
ParameterSet.hasDefaultOrIsSet()-method depends on the value of
this Parameter.
- getNeededReference() -
Method in class de.jstacs.parameters.RangeParameter
-
- getNeededReferenceId() -
Method in class de.jstacs.parameters.Parameter
- Returns the id of the
ParameterSet that would be returned by
Parameter.getNeededReference().
- getNewAlphabet() -
Method in class de.jstacs.data.alphabets.DiscreteAlphabetMapping
- Returns the new Alphabet that is used for mapping.
- getNewAlphabetContainer(AlphabetContainer, DiscreteAlphabetMapping...) -
Static method in class de.jstacs.data.sequences.MappedDiscreteSequence
- This method allows to create a new
AlphabetContainer given an old AlphabetContainer and some DiscreteAlphabetMappings.
- getNewComponentProbs(double[]) -
Method in class de.jstacs.models.mixture.AbstractMixtureModel
- Estimates the weights of each component.
- getNewDiscreteValue(int) -
Method in class de.jstacs.data.alphabets.DiscreteAlphabetMapping
- This method implements the main transformation of the values.
- getNewInstance() -
Method in class de.jstacs.classifier.scoringFunctionBased.logPrior.CompositeLogPrior
-
- getNewInstance() -
Method in class de.jstacs.classifier.scoringFunctionBased.logPrior.DoesNothingLogPrior
-
- getNewInstance() -
Method in class de.jstacs.classifier.scoringFunctionBased.logPrior.LogPrior
- This method returns an empty new instance of the current prior.
- getNewInstance() -
Method in class de.jstacs.classifier.scoringFunctionBased.logPrior.SeparateLogPrior
-
- getNewInstance() -
Method in class de.jstacs.classifier.scoringFunctionBased.logPrior.SimpleGaussianSumLogPrior
-
- getNewParameters(int, double[][], double[]) -
Method in class de.jstacs.models.discrete.inhomogeneous.shared.SharedStructureMixture
-
- getNewParameters() -
Method in class de.jstacs.models.hmm.models.SamplingHigherOrderHMM
- This method set all parameters for the next sampling step
- getNewParameters() -
Method in class de.jstacs.models.hmm.models.SamplingPhyloHMM
-
- getNewParameters(int, double[][], double[]) -
Method in class de.jstacs.models.mixture.AbstractMixtureModel
- This method trains the internal models on the internal sample and the
given weights.
- getNewParameters(int, double[][], double[]) -
Method in class de.jstacs.models.mixture.motif.HiddenMotifMixture
-
- getNewParametersForModel(int, int, int, double[]) -
Method in class de.jstacs.models.mixture.AbstractMixtureModel
- This method trains the internal model with index
modelIndex
on the internal sample and the given weights.
- getNewStartDistance() -
Method in class de.jstacs.algorithms.optimization.ConstantStartDistance
-
- getNewStartDistance() -
Method in class de.jstacs.algorithms.optimization.LimitedMedianStartDistance
-
- getNewStartDistance() -
Method in interface de.jstacs.algorithms.optimization.StartDistanceForecaster
- This method returns the new positive start distance.
- getNewWeights(double[], double[], double[][]) -
Method in class de.jstacs.models.mixture.AbstractMixtureModel
- Computes sequence weights and returns the score.
- getNewWeights(double[], double[], double[][]) -
Method in class de.jstacs.models.mixture.MixtureModel
- Computes sequence weights and returns the score.
- getNewWeights(double[], double[], double[][]) -
Method in class de.jstacs.models.mixture.motif.SingleHiddenMotifMixture
-
- getNewWeights(double[], double[], double[][]) -
Method in class de.jstacs.models.mixture.StrandModel
- Computes sequence weights and returns the score.
- getNextContext(int, int) -
Method in class de.jstacs.models.hmm.transitions.BasicHigherOrderTransition.AbstractTransitionElement
- This method returns the next context that will be visited when visiting the child with index
index.
- getNodeLabel(double, String, NumberFormat) -
Method in class de.jstacs.models.hmm.states.emissions.continuous.GaussianEmission
-
- getNodeLabel(double, String, NumberFormat) -
Method in class de.jstacs.models.hmm.states.emissions.discrete.AbstractConditionalDiscreteEmission
-
- getNodeLabel(double, String, NumberFormat) -
Method in interface de.jstacs.models.hmm.states.emissions.Emission
- Returns the graphviz label of the node containing this emission.
- getNodeLabel(double, String, NumberFormat) -
Method in class de.jstacs.models.hmm.states.emissions.MixtureEmission
-
- getNodeLabel(double, String, NumberFormat) -
Method in class de.jstacs.models.hmm.states.emissions.SilentEmission
-
- getNodeShape(boolean) -
Method in class de.jstacs.models.hmm.states.emissions.continuous.GaussianEmission
-
- getNodeShape(boolean) -
Method in class de.jstacs.models.hmm.states.emissions.discrete.AbstractConditionalDiscreteEmission
-
- getNodeShape(boolean) -
Method in interface de.jstacs.models.hmm.states.emissions.Emission
- Returns the graphviz string for the shape of the node.
- getNodeShape(boolean) -
Method in class de.jstacs.models.hmm.states.emissions.MixtureEmission
-
- getNodeShape(boolean) -
Method in class de.jstacs.models.hmm.states.emissions.SilentEmission
-
- getNormalizedVersion(NormalizableScoringFunction, int) -
Static method in class de.jstacs.scoringFunctions.NormalizedScoringFunction
- This method returns a normalized version of a NormalizableScoringFunction.
- getNucleicAcid() -
Method in enum de.jstacs.data.DinucleotideProperty
- Returns the kind of nucleic acid, e.g.
- getNumberOfAlignedSequences() -
Method in class de.jstacs.algorithms.alignment.StringAlignment
- Returns the number of sequences in this alignment.
- getNumberOfAllNodesBelow() -
Method in class de.jstacs.models.phylo.PhyloNode
- This method returns the total number of
PhyloNodes in the subtree starting from this instance
- getNumberOfAlphabets() -
Method in class de.jstacs.data.AlphabetContainer
- This method returns the number of
Alphabets used in the current AlphabetContainer.
- getNumberOfAvailableProcessors() -
Static method in class de.jstacs.classifier.scoringFunctionBased.AbstractMultiThreadedOptimizableFunction
- This method returns the number of available processors.
- getNumberOfBoundSequences(Sample, double[], int) -
Method in class de.jstacs.motifDiscovery.SignificantMotifOccurrencesFinder
- Returns the number of sequences in
data that are predicted to be bound at least once by motif no.
- getNumberOfChildren(int, int) -
Method in interface de.jstacs.models.hmm.Transition
- This method returns the number of children states for given index, i.e. context, and
a given layer of the matrix.
- getNumberOfChildren() -
Method in class de.jstacs.models.hmm.transitions.BasicHigherOrderTransition.AbstractTransitionElement
- This method returns the number of states that can be visited.
- getNumberOfChildren(int, int) -
Method in class de.jstacs.models.hmm.transitions.BasicHigherOrderTransition
-
- getNumberOfClasses() -
Method in class de.jstacs.classifier.AbstractClassifier
- Returns the number of classes that can be distinguished.
- getNumberOfClasses() -
Method in class de.jstacs.classifier.AbstractScoreBasedClassifier
-
- getNumberOfCombinations(int) -
Method in class de.jstacs.models.discrete.inhomogeneous.CombinationIterator
- Returns the number of possible combinations.
- getNumberOfComponents() -
Method in class de.jstacs.models.mixture.AbstractMixtureModel
- Returns the number of components the are modeled by this
AbstractMixtureModel.
- getNumberOfComponents() -
Method in interface de.jstacs.motifDiscovery.MotifDiscoverer
- Returns the number of components in this
MotifDiscoverer.
- getNumberOfComponents() -
Method in class de.jstacs.scoringFunctions.IndependentProductScoringFunction
-
- getNumberOfComponents() -
Method in class de.jstacs.scoringFunctions.MappingScoringFunction
-
- getNumberOfComponents() -
Method in class de.jstacs.scoringFunctions.mix.AbstractMixtureScoringFunction
- Returns the number of different components of this
AbstractMixtureScoringFunction.
- getNumberOfElements() -
Method in class de.jstacs.data.Sample
- Returns the number of elements, i.e. the
Sequences, in this
Sample.
- getNumberOfElements() -
Method in class de.jstacs.data.Sample.WeightedSampleFactory
- Returns the number of elements, i.e.
- getNumberOfElements() -
Method in class de.jstacs.io.StringExtractor
- Returns the number of
Strings that have been read.
- getNumberOfElementsWithLength(int) -
Method in class de.jstacs.data.Sample
- Returns the number of overlapping elements that can be extracted.
- getNumberOfElementsWithLength(int, double[]) -
Method in class de.jstacs.data.Sample
- Returns the weighted number of overlapping elements that can be extracted.
- getNumberOfIndexes(int) -
Method in interface de.jstacs.models.hmm.Transition
- This method computes the number of different indexes for a given layer of the matrix.
- getNumberOfIndexes(int) -
Method in class de.jstacs.models.hmm.transitions.BasicHigherOrderTransition
-
- getNumberOfLines() -
Method in class de.jstacs.classifier.AbstractScoreBasedClassifier.DoubleTableResult
- Returns the number of lines in this table.
- getNumberOfModels() -
Method in class de.jstacs.models.CompositeModel
- This method returns the number of models in the
CompositeModel.
- getNumberOfMotifs() -
Method in class de.jstacs.models.mixture.motif.SingleHiddenMotifMixture
-
- getNumberOfMotifs() -
Method in interface de.jstacs.motifDiscovery.MotifDiscoverer
- Returns the number of motifs for this
MotifDiscoverer.
- getNumberOfMotifs() -
Method in class de.jstacs.scoringFunctions.IndependentProductScoringFunction
-
- getNumberOfMotifs() -
Method in class de.jstacs.scoringFunctions.MappingScoringFunction
-
- getNumberOfMotifs() -
Method in class de.jstacs.scoringFunctions.mix.MixtureScoringFunction
-
- getNumberOfMotifs() -
Method in class de.jstacs.scoringFunctions.mix.motifSearch.HiddenMotifsMixture
-
- getNumberOfMotifsInComponent(int) -
Method in class de.jstacs.models.mixture.motif.SingleHiddenMotifMixture
-
- getNumberOfMotifsInComponent(int) -
Method in interface de.jstacs.motifDiscovery.MotifDiscoverer
- Returns the number of motifs that are used in the component
component of this MotifDiscoverer.
- getNumberOfMotifsInComponent(int) -
Method in class de.jstacs.scoringFunctions.IndependentProductScoringFunction
-
- getNumberOfMotifsInComponent(int) -
Method in class de.jstacs.scoringFunctions.MappingScoringFunction
-
- getNumberOfMotifsInComponent(int) -
Method in class de.jstacs.scoringFunctions.mix.MixtureScoringFunction
-
- getNumberOfMotifsInComponent(int) -
Method in class de.jstacs.scoringFunctions.mix.motifSearch.HiddenMotifsMixture
-
- getNumberOfNexts(int) -
Method in class de.jstacs.parameters.MultiSelectionCollectionParameter
- Returns the number of calls of
MultiSelectionCollectionParameter.next() that can be called
before false is returned.
- getNumberOfNexts(int) -
Method in class de.jstacs.parameters.RangeParameter
- Returns the number of calls of
RangeParameter.next() that can be done before
obtaining false.
- getNumberOfNodes() -
Method in class de.jstacs.algorithms.graphs.tensor.Tensor
- Returns the number of nodes.
- getNumberOfNodes() -
Method in class de.jstacs.models.phylo.PhyloTree
- This method returns the total number of nodes in the tree
- getNumberOfParameters() -
Method in class de.jstacs.models.hmm.models.DifferentiableHigherOrderHMM
-
- getNumberOfParameters() -
Method in class de.jstacs.models.hmm.states.emissions.continuous.GaussianEmission
-
- getNumberOfParameters() -
Method in interface de.jstacs.models.hmm.states.emissions.DifferentiableEmission
- Returns the number of parameters of this emission.
- getNumberOfParameters() -
Method in class de.jstacs.models.hmm.states.emissions.discrete.AbstractConditionalDiscreteEmission
-
- getNumberOfParameters() -
Method in class de.jstacs.models.hmm.states.emissions.SilentEmission
-
- getNumberOfParameters() -
Method in class de.jstacs.models.hmm.transitions.BasicHigherOrderTransition.AbstractTransitionElement
- This method returns the number of parameters in this transition element.
- getNumberOfParameters() -
Method in class de.jstacs.parameters.ArrayParameterSet
-
- getNumberOfParameters() -
Method in class de.jstacs.parameters.ParameterSet
- Returns the number of parameters in the
ParameterSet.
- getNumberOfParameters() -
Method in class de.jstacs.parameters.SequenceScoringParameterSet
-
- getNumberOfParameters() -
Method in class de.jstacs.scoringFunctions.CMMScoringFunction
-
- getNumberOfParameters() -
Method in class de.jstacs.scoringFunctions.directedGraphicalModels.BayesianNetworkScoringFunction
-
- getNumberOfParameters() -
Method in class de.jstacs.scoringFunctions.homogeneous.HMM0ScoringFunction
-
- getNumberOfParameters() -
Method in class de.jstacs.scoringFunctions.homogeneous.HMMScoringFunction
-
- getNumberOfParameters() -
Method in class de.jstacs.scoringFunctions.homogeneous.UniformHomogeneousScoringFunction
-
- getNumberOfParameters() -
Method in class de.jstacs.scoringFunctions.IndependentProductScoringFunction
-
- getNumberOfParameters() -
Method in class de.jstacs.scoringFunctions.MappingScoringFunction
-
- getNumberOfParameters() -
Method in class de.jstacs.scoringFunctions.mix.AbstractMixtureScoringFunction
-
- getNumberOfParameters() -
Method in class de.jstacs.scoringFunctions.mix.motifSearch.MixtureDuration
-
- getNumberOfParameters() -
Method in class de.jstacs.scoringFunctions.mix.motifSearch.SkewNormalLikeScoringFunction
-
- getNumberOfParameters() -
Method in class de.jstacs.scoringFunctions.mix.motifSearch.UniformDurationScoringFunction
-
- getNumberOfParameters() -
Method in class de.jstacs.scoringFunctions.MRFScoringFunction
-
- getNumberOfParameters() -
Method in class de.jstacs.scoringFunctions.NormalizedScoringFunction
-
- getNumberOfParameters() -
Method in interface de.jstacs.scoringFunctions.ScoringFunction
- Returns the number of parameters in this
ScoringFunction.
- getNumberOfParameters() -
Method in class de.jstacs.scoringFunctions.UniformScoringFunction
-
- getNumberOfParents() -
Method in class de.jstacs.scoringFunctions.directedGraphicalModels.ParameterTree
- Returns the number of parents for the random variable of this
ParameterTree in the network structure of the enclosing
BayesianNetworkScoringFunction.
- getNumberOfPossibilities() -
Method in class de.jstacs.scoringFunctions.mix.motifSearch.DurationScoringFunction
- Returns the number of different possibilities that can be scored.
- getNumberOfRecommendedStarts() -
Method in class de.jstacs.models.hmm.models.DifferentiableHigherOrderHMM
-
- getNumberOfRecommendedStarts() -
Method in class de.jstacs.scoringFunctions.AbstractNormalizableScoringFunction
-
- getNumberOfRecommendedStarts() -
Method in class de.jstacs.scoringFunctions.CMMScoringFunction
-
- getNumberOfRecommendedStarts() -
Method in class de.jstacs.scoringFunctions.homogeneous.HMMScoringFunction
-
- getNumberOfRecommendedStarts() -
Method in class de.jstacs.scoringFunctions.IndependentProductScoringFunction
-
- getNumberOfRecommendedStarts() -
Method in class de.jstacs.scoringFunctions.mix.AbstractMixtureScoringFunction
-
- getNumberOfRecommendedStarts() -
Method in class de.jstacs.scoringFunctions.mix.motifSearch.MixtureDuration
-
- getNumberOfRecommendedStarts() -
Method in class de.jstacs.scoringFunctions.mix.motifSearch.SkewNormalLikeScoringFunction
-
- getNumberOfRecommendedStarts() -
Method in class de.jstacs.scoringFunctions.NormalizedScoringFunction
-
- getNumberOfRecommendedStarts() -
Method in interface de.jstacs.scoringFunctions.ScoringFunction
- This method returns the number of recommended optimization starts.
- getNumberOfResults() -
Method in class de.jstacs.results.ResultSet
- Returns the number of
Results in this ResultSet
- getNumberOfSequenceAnnotationsByType(String) -
Method in class de.jstacs.data.Sequence
- Returns the number of
SequenceAnnotations of type type for this Sequence.
- getNumberOfSequences() -
Method in class de.jstacs.models.discrete.inhomogeneous.SequenceIterator
- This method returns the number of sequences in this iterator,
i.e., the number of times
SequenceIterator.next() returns true after using SequenceIterator.reset().
- getNumberOfSpecificConstraints() -
Method in class de.jstacs.models.discrete.Constraint
- Returns the number of specific constraints.
- getNumberOfStarts() -
Method in class de.jstacs.classifier.scoringFunctionBased.sampling.SamplingScoreBasedClassifierParameterSet
- Returns the number of independent sampling starts
- getNumberOfStarts() -
Method in class de.jstacs.models.hmm.HMMTrainingParameterSet
- The method returns the number of different starts.
- getNumberOfStarts() -
Method in class de.jstacs.sampling.AbstractBurnInTestParameterSet
- Returns the number of starts.
- getNumberOfStarts(ScoringFunction[]) -
Static method in class de.jstacs.scoringFunctions.AbstractNormalizableScoringFunction
- Returns the number of recommended starts in a numerical optimization.
- getNumberOfStates() -
Method in class de.jstacs.models.hmm.AbstractHMM
- This method returns the number of the (hidden) states
- getNumberOfStates() -
Method in interface de.jstacs.models.hmm.Transition
- This method returns the number of states underlying this transition
instance.
- getNumberOfStates() -
Method in class de.jstacs.models.hmm.transitions.BasicHigherOrderTransition
-
- getNumberOfStationarySamplings() -
Method in class de.jstacs.classifier.scoringFunctionBased.sampling.SamplingScoreBasedClassifierParameterSet
- Returns the number of samplings steps in the stationary phase
- getNumberOfStepsInStationaryPhase() -
Method in class de.jstacs.models.hmm.training.SamplingHMMTrainingParameterSet
- The method returns the number of steps to be done in the stationary phase.
- getNumberOfStepsPerIteration() -
Method in class de.jstacs.models.hmm.training.SamplingHMMTrainingParameterSet
- This method returns the number of steps to be done in each start before testing for the end of the burn in phase (again).
- getNumberOfTestSamplings() -
Method in class de.jstacs.classifier.scoringFunctionBased.sampling.SamplingScoreBasedClassifierParameterSet
- Returns the number of samplings between checks for the stationary phase
- getNumberOfThreads() -
Method in class de.jstacs.classifier.scoringFunctionBased.AbstractMultiThreadedOptimizableFunction
- Returns the number of used threads for evaluating the function and for determining the gradient of the function.
- getNumberOfThreads() -
Method in class de.jstacs.classifier.scoringFunctionBased.gendismix.GenDisMixClassifier
- This method returns the number of used threads while optimization.
- getNumberOfThreads() -
Method in class de.jstacs.classifier.scoringFunctionBased.gendismix.GenDisMixClassifierParameterSet
- This method returns the number of threads that should be used during optimization.
- getNumberOfThreads() -
Method in class de.jstacs.classifier.scoringFunctionBased.sampling.SamplingGenDisMixClassifierParameterSet
- Returns the number of threads for evaluating the
LogGenDisMixFunction
- getNumberOfThreads() -
Method in class de.jstacs.models.hmm.training.NumericalHMMTrainingParameterSet
- This method returns the number of threads that should be used during optimization.
- getNumberOfValues() -
Method in class de.jstacs.parameters.MultiSelectionCollectionParameter
-
- getNumberOfValues() -
Method in class de.jstacs.parameters.ParameterSet
-
- getNumberOfValues() -
Method in class de.jstacs.parameters.ParameterSetContainer
-
- getNumberOfValues() -
Method in interface de.jstacs.parameters.RangeIterator
- Returns the number of values in the collection.
- getNumberOfValues() -
Method in class de.jstacs.parameters.RangeParameter
- Returns the number of values in a list or range of parameter values.
- getNumericalCharacteristics() -
Method in class de.jstacs.classifier.AbstractClassifier
- Returns the subset of numerical values that are also returned by
AbstractClassifier.getCharacteristics().
- getNumericalCharacteristics() -
Method in class de.jstacs.classifier.MappingClassifier
-
- getNumericalCharacteristics() -
Method in class de.jstacs.classifier.modelBased.ModelBasedClassifier
-
- getNumericalCharacteristics() -
Method in class de.jstacs.classifier.scoringFunctionBased.sampling.SamplingScoreBasedClassifier
-
- getNumericalCharacteristics() -
Method in class de.jstacs.classifier.scoringFunctionBased.ScoreClassifier
-
- getNumericalCharacteristics() -
Method in class de.jstacs.models.CompositeModel
-
- getNumericalCharacteristics() -
Method in class de.jstacs.models.discrete.homogeneous.HomogeneousModel
-
- getNumericalCharacteristics() -
Method in class de.jstacs.models.discrete.inhomogeneous.DAGModel
-
- getNumericalCharacteristics() -
Method in class de.jstacs.models.hmm.models.HigherOrderHMM
-
- getNumericalCharacteristics() -
Method in class de.jstacs.models.mixture.AbstractMixtureModel
-
- getNumericalCharacteristics() -
Method in interface de.jstacs.models.Model
- Returns the subset of numerical values that are also returned by
Model.getCharacteristics().
- getNumericalCharacteristics() -
Method in class de.jstacs.models.NormalizableScoringFunctionModel
-
- getNumericalCharacteristics() -
Method in class de.jstacs.models.UniformModel
-
- getNumericalCharacteristics() -
Method in class de.jstacs.models.VariableLengthWrapperModel
-
- getOffsetForAucPR() -
Method in class de.jstacs.motifDiscovery.SignificantMotifOccurrencesFinder
- This method returns an offset that must be added to scores for computing PR curves.
- getOptimalBranching(double[][], double[][], byte) -
Static method in class de.jstacs.algorithms.graphs.Chu_Liu_Edmonds
- Returns an optimal branching.
- getOrder() -
Method in class de.jstacs.algorithms.graphs.tensor.Tensor
- Returns the order.
- getOrder() -
Method in enum de.jstacs.motifDiscovery.SignificantMotifOccurrencesFinder.RandomSeqType
- This method returns the Markov order.
- getOrder() -
Method in class de.jstacs.scoringFunctions.directedGraphicalModels.MutableMarkovModelScoringFunction
- Returns the order of the inhomogeneous Markov model.
- getOrder() -
Method in class de.jstacs.scoringFunctions.directedGraphicalModels.structureLearning.measures.InhomogeneousMarkov
- Returns the order of the Markov model as defined in the constructor
- getOrder() -
Method in class de.jstacs.scoringFunctions.directedGraphicalModels.structureLearning.measures.InhomogeneousMarkov.InhomogeneousMarkovParameterSet
- Returns the order of the
InhomogeneousMarkov structure
measure as defined by this set of parameters.
- getOrder() -
Method in class de.jstacs.scoringFunctions.directedGraphicalModels.structureLearning.measures.pmmMeasures.PMMExplainingAwayResidual.PMMExplainingAwayResidualParameterSet
- Returns the order defined by this set of parameters.
- getOrder() -
Method in class de.jstacs.scoringFunctions.directedGraphicalModels.structureLearning.measures.pmmMeasures.PMMMutualInformation.PMMMutualInformationParameterSet
- Returns the order defined by this set of parameters.
- getOutfilePrefix() -
Method in class de.jstacs.classifier.scoringFunctionBased.sampling.SamplingScoreBasedClassifierParameterSet
- Returns the prefix of the temporary files for storing sampled
parameter values
- getOutput(byte[], double) -
Method in class de.jstacs.models.discrete.inhomogeneous.InhCondProb
- This method is used to create random sequences.
- getOutputStream() -
Method in class de.jstacs.utils.galaxy.GalaxyAdaptor.Protocol
- Returns the
ByteArrayOutputStream of this protocol
- getOutputStream() -
Method in class de.jstacs.utils.SafeOutputStream
- Returns the internal used
OutputStream.
- getParameterAt(int) -
Method in class de.jstacs.parameters.ArrayParameterSet
-
- getParameterAt(int) -
Method in class de.jstacs.parameters.ParameterSet
- Returns the
Parameter at position i.
- getParameterAt(int) -
Method in class de.jstacs.parameters.SequenceScoringParameterSet
-
- getParameterFor(Sequence, int) -
Method in class de.jstacs.scoringFunctions.directedGraphicalModels.ParameterTree
- Returns the
Parameter that is responsible for the suffix of
sequence seq starting at position start.
- getParameterFromTag(String) -
Method in class de.jstacs.parameters.ParameterSetTagger
- This method returns the
Parameter specified by the tag
- getParameterIndexesForSamplingStep(int, int) -
Method in class de.jstacs.scoringFunctions.directedGraphicalModels.ParameterTree
- Returns the indexes of the parameters, incremented by
offset, that
shall be sampled in step step of a grouped sampling process.
- getParameters(OptimizableFunction.KindOfParameter, double[]) -
Method in class de.jstacs.classifier.scoringFunctionBased.AbstractOptimizableFunction
- This method enables the user to get the parameters without creating a new
array.
- getParameters(OptimizableFunction.KindOfParameter) -
Method in class de.jstacs.classifier.scoringFunctionBased.AbstractOptimizableFunction
-
- getParameters(OptimizableFunction.KindOfParameter) -
Method in class de.jstacs.classifier.scoringFunctionBased.OptimizableFunction
- Returns some parameters that can be used for instance as start
parameters.
- getParameters(OptimizableFunction.KindOfParameter, double[]) -
Method in class de.jstacs.classifier.scoringFunctionBased.SFBasedOptimizableFunction
-
- getParameterSetFor(Class<? extends InstantiableFromParameterSet>) -
Static method in class de.jstacs.utils.SubclassFinder
- Returns a
LinkedList of the classes of the
InstanceParameterSets that can be used to instantiate the
sub-class of InstantiableFromParameterSet that is given by
clazz
- getParametersInCollection() -
Method in class de.jstacs.parameters.CollectionParameter
- Returns the possible values in this collection.
- getParent() -
Method in class de.jstacs.parameters.Parameter
- Returns a reference to the
ParameterSet enclosing this
Parameter.
- getParent() -
Method in class de.jstacs.parameters.ParameterSet
- Returns the enclosing
ParameterSetContainer of this
ParameterSet or null if none exists.
- getParents(Sample, Sample, double[], double[], int) -
Method in class de.jstacs.scoringFunctions.directedGraphicalModels.structureLearning.measures.btMeasures.BTExplainingAwayResidual
-
- getParents(Sample, Sample, double[], double[], int) -
Method in class de.jstacs.scoringFunctions.directedGraphicalModels.structureLearning.measures.btMeasures.BTMutualInformation
-
- getParents(Sample, Sample, double[], double[], int) -
Method in class de.jstacs.scoringFunctions.directedGraphicalModels.structureLearning.measures.InhomogeneousMarkov
-
- getParents(Sample, Sample, double[], double[], int) -
Method in class de.jstacs.scoringFunctions.directedGraphicalModels.structureLearning.measures.Measure
- Returns the optimal parents for the given data and weights.
- getParents(Sample, Sample, double[], double[], int) -
Method in class de.jstacs.scoringFunctions.directedGraphicalModels.structureLearning.measures.pmmMeasures.PMMExplainingAwayResidual
-
- getParents(Sample, Sample, double[], double[], int) -
Method in class de.jstacs.scoringFunctions.directedGraphicalModels.structureLearning.measures.pmmMeasures.PMMMutualInformation
-
- getParser() -
Method in class de.jstacs.results.SampleResult
- Returns the
SequenceAnnotationParser that can be used to
write this SampleResult including annotations on the contained Sequences
to a file.
- getPartialLengths() -
Method in class de.jstacs.scoringFunctions.IndependentProductScoringFunction
- This method returns a deep copy of the internally used partial lengths of the parts.
- getPartialROC(double[], double[], RangeParameter) -
Static method in class de.jstacs.classifier.ScoreBasedPerformanceMeasureDefinitions
- This method allows to compute a partial ROC curve.
- getPath() -
Method in class de.jstacs.utils.galaxy.GalaxyAdaptor.FileResult
- Returns the path of the directory containing the file
- getPercent() -
Method in class de.jstacs.classifier.assessment.Sampled_RepeatedHoldOutAssessParameterSet
- Returns the percentage of user supplied data that is used in each
iteration as test dataset.
- getPercents() -
Method in class de.jstacs.classifier.assessment.RepeatedHoldOutAssessParameterSet
- Returns an array containing for each class the percentage of user
supplied data that is used in each iteration as test dataset.
- getPlotCommands(REnvironment, String, AbstractScoreBasedClassifier.DoubleTableResult...) -
Static method in class de.jstacs.classifier.AbstractScoreBasedClassifier.DoubleTableResult
- This method copies the data to the server side and creates a
StringBuffer containing the plot commands.
- getPlotCommands(REnvironment, String, int[], AbstractScoreBasedClassifier.DoubleTableResult...) -
Static method in class de.jstacs.classifier.AbstractScoreBasedClassifier.DoubleTableResult
- This method copies the data to the server side and creates a
StringBuffer containing the plot commands.
- getPlotCommands(REnvironment, String, String[], AbstractScoreBasedClassifier.DoubleTableResult...) -
Static method in class de.jstacs.classifier.AbstractScoreBasedClassifier.DoubleTableResult
- This method copies the data to the server side and creates a
StringBuffer containing the plot commands.
- getPlugInParameters() -
Method in class de.jstacs.scoringFunctions.directedGraphicalModels.BayesianNetworkScoringFunctionParameterSet
- Returns true if plug-in parameters shall be used when creating a
BayesianNetworkScoringFunction from this set of parameters.
- getPosition() -
Method in class de.jstacs.data.sequences.annotation.LocatedSequenceAnnotation
- Returns the position of this
LocatedSequenceAnnotation on the
sequence.
- getPosition(int) -
Method in class de.jstacs.models.discrete.Constraint
- Returns the position with index
index.
- getPosition() -
Method in class de.jstacs.scoringFunctions.directedGraphicalModels.Parameter
- Returns the position of the symbol this parameter is responsible for as
defined in the constructor.
- getPositionDependentKMerProb(Sequence) -
Method in class de.jstacs.scoringFunctions.directedGraphicalModels.BayesianNetworkScoringFunction
- Returns the probability of
kmer for all possible positions in this BayesianNetworkScoringFunction starting at position kmer.getLength()-1.
- getPositionForParameter(int) -
Method in class de.jstacs.scoringFunctions.directedGraphicalModels.BayesianNetworkScoringFunction
- Returns the position in the sequence the parameter
index is
responsible for.
- getPositions() -
Method in class de.jstacs.models.discrete.Constraint
- Returns a clone of the array of used positions.
- getPossibleLength(Model...) -
Static method in class de.jstacs.classifier.modelBased.ModelBasedClassifier
- This method returns the possible length of a classifier that would use
the given
Models.
- getPossibleLength() -
Method in class de.jstacs.data.AlphabetContainer
- Returns the possible length for
Sequences using this
AlphabetContainer.
- getPossibleLength() -
Method in class de.jstacs.data.AlphabetContainerParameterSet
- Returns the length of the
AlphabetContainer that can be instantiated using
this AlphabetContainerParameterSet.SectionDefinedAlphabetParameterSet.
- getPPVForSensitivity(double[], double[], double) -
Static method in class de.jstacs.classifier.ScoreBasedPerformanceMeasureDefinitions
- This method computes the positive predictive value (PPV) for a given
sensitivity.
- getPriorTerm() -
Method in class de.jstacs.models.AbstractModel
-
- getPriorTerm() -
Method in class de.jstacs.models.hmm.AbstractHMM
-
- getPriorTerm() -
Method in interface de.jstacs.models.Model
- Returns a value that is proportional to the prior.
- getProbFor(Sequence) -
Method in class de.jstacs.models.AbstractModel
-
- getProbFor(Sequence, int) -
Method in class de.jstacs.models.AbstractModel
-
- getProbFor(Sequence, int, int) -
Method in class de.jstacs.models.CompositeModel
-
- getProbFor(Sequence, int, int) -
Method in class de.jstacs.models.discrete.homogeneous.HomogeneousModel
-
- getProbFor(Sequence, int, int) -
Method in class de.jstacs.models.discrete.inhomogeneous.DAGModel
-
- getProbFor(Sequence) -
Method in class de.jstacs.models.hmm.AbstractHMM
-
- getProbFor(Sequence, int) -
Method in class de.jstacs.models.hmm.AbstractHMM
-
- getProbFor(Sequence, int, int) -
Method in class de.jstacs.models.hmm.AbstractHMM
-
- getProbFor(Sequence, int, int) -
Method in class de.jstacs.models.mixture.AbstractMixtureModel
-
- getProbFor(Sequence) -
Method in interface de.jstacs.models.Model
- Returns the probability of the given sequence given the model.
- getProbFor(Sequence, int) -
Method in interface de.jstacs.models.Model
- Returns the probability of (a part of) the given sequence given the
model.
- getProbFor(Sequence, int, int) -
Method in interface de.jstacs.models.Model
- Returns the probability of (a part of) the given sequence given the
model.
- getProbFor(Sequence, int, int) -
Method in class de.jstacs.models.NormalizableScoringFunctionModel
-
- getProbFor(Sequence, int, int) -
Method in class de.jstacs.models.UniformModel
-
- getProbFor(Sequence, int, int) -
Method in class de.jstacs.models.VariableLengthWrapperModel
-
- getProbFor(Sequence) -
Method in class de.jstacs.scoringFunctions.directedGraphicalModels.ParameterTree
- Returns the probability of
Sequence sequence in this ParameterTree.
- getProbsForComponent(Sequence) -
Method in class de.jstacs.scoringFunctions.mix.AbstractMixtureScoringFunction
- Returns the probabilities for each component given a
Sequence.
- getProfileOfScoresFor(int, int, Sequence, int, MotifDiscoverer.KindOfProfile) -
Method in class de.jstacs.models.mixture.motif.SingleHiddenMotifMixture
-
- getProfileOfScoresFor(int, int, Sequence, int, MotifDiscoverer.KindOfProfile) -
Method in interface de.jstacs.motifDiscovery.MotifDiscoverer
- Returns the profile of the scores for component
component
and motif motif at all possible start positions of the motif
in the sequence sequence beginning at startpos.
- getProfileOfScoresFor(int, int, Sequence, int, MotifDiscoverer.KindOfProfile) -
Method in class de.jstacs.scoringFunctions.IndependentProductScoringFunction
-
- getProfileOfScoresFor(int, int, Sequence, int, MotifDiscoverer.KindOfProfile) -
Method in class de.jstacs.scoringFunctions.MappingScoringFunction
-
- getProfileOfScoresFor(int, int, Sequence, int, MotifDiscoverer.KindOfProfile) -
Method in class de.jstacs.scoringFunctions.mix.MixtureScoringFunction
-
- getProfileOfScoresFor(int, int, Sequence, int, MotifDiscoverer.KindOfProfile) -
Method in class de.jstacs.scoringFunctions.mix.motifSearch.HiddenMotifsMixture
-
- getProperty(Sequence, DinucleotideProperty.Smoothing) -
Method in enum de.jstacs.data.DinucleotideProperty
- Computes this dinucleotide property for all overlapping twomers in
original, smoothes the result using smoothing,
and returns the smoothed property as a double array.
- getProperty(Sequence) -
Method in enum de.jstacs.data.DinucleotideProperty
- Computes this dinucleotide property for all overlapping twomers in
original
and returns the result as a double array of length original.getLength()-1
- getPropertyAsSequence(Sequence) -
Method in enum de.jstacs.data.DinucleotideProperty
- Computes this dinucleotide property for all overlapping twomers in
original
and returns the result as a Sequence of length original.getLength()-1
- getPropertyAsSequence(Sequence, DinucleotideProperty.Smoothing) -
Method in enum de.jstacs.data.DinucleotideProperty
- Computes this dinucleotide property for all overlapping twomers in
original, smoothes the result using smoothing,
and returns the smoothed property as a Sequence.
- getPropertyImage(Sequence, DinucleotideProperty, DinucleotideProperty.Smoothing, REnvironment, int, String, int, int) -
Static method in enum de.jstacs.data.DinucleotideProperty
-
- getPropertyImage(Sample, DinucleotideProperty, DinucleotideProperty.Smoothing, REnvironment, int, String, int, int) -
Static method in enum de.jstacs.data.DinucleotideProperty
-
- getProtocol(boolean) -
Method in class de.jstacs.utils.galaxy.GalaxyAdaptor
- Returns an object for writing a protocol of a program run
- getPseudoCount() -
Method in class de.jstacs.scoringFunctions.directedGraphicalModels.Parameter
- Returns the pseudocount as given in the constructor.
- getPubMedID() -
Method in enum de.jstacs.data.DinucleotideProperty
- Returns the PubMed ID of the publication where the parameters of this property has been published.
- getPValue(Sequence, Sample) -
Method in class de.jstacs.classifier.AbstractScoreBasedClassifier
- Returns the p-value for a
Sequence candidate with
respect to a given background Sample.
- getPValue(Sample, Sample) -
Method in class de.jstacs.classifier.AbstractScoreBasedClassifier
- Returns the p-values for all
Sequences in the Sample
candidates with respect to a given background Sample
.
- getPValue(double[], double) -
Static method in class de.jstacs.classifier.utils.PValueComputation
- This method searches for the insertion point of the score in a given
sorted array of scores and returns the p-value for this score.
- getPValue(double[], double, int) -
Static method in class de.jstacs.classifier.utils.PValueComputation
- This method searches for the insertion point of the score in a given
sorted array of scores from index
start and returns the
p-value for this score.
- getPWM() -
Method in class de.jstacs.scoringFunctions.directedGraphicalModels.BayesianNetworkScoringFunction
- If this
BayesianNetworkScoringFunction is a PWM, i.e.
- getRandomSequence(Random, int) -
Method in class de.jstacs.models.discrete.homogeneous.HomogeneousMM
-
- getRandomSequence(Random, int) -
Method in class de.jstacs.models.discrete.homogeneous.HomogeneousModel
- This method creates a random
Sequence from a trained homogeneous
model.
- getRangedInstance() -
Method in class de.jstacs.parameters.CollectionParameter
-
- getRangedInstance() -
Method in class de.jstacs.parameters.ParameterSetContainer
-
- getRangedInstance() -
Method in interface de.jstacs.parameters.Rangeable
- Returns an instance of
RangeIterator that has the same properties
as the current instance, but accepts a range or list of values.
- getRangedInstance() -
Method in class de.jstacs.parameters.SimpleParameter
-
- getRawResult() -
Method in class de.jstacs.results.ListResult
- Returns a copy of the internal list of
ResultSets.
- getReference() -
Method in enum de.jstacs.data.DinucleotideProperty
- Returns the reference of the publication where the parameters of this property has been published.
- getReferenceClass() -
Method in class de.jstacs.classifier.assessment.Sampled_RepeatedHoldOutAssessParameterSet
- Returns the index of the reference class.
- getReferenceSequence() -
Method in class de.jstacs.data.sequences.annotation.ReferenceSequenceAnnotation
- Returns the reference sequence.
- getRegEx() -
Method in class de.jstacs.io.RegExFilenameFilter
- Returns a representation of all used regular expressions.
- getRepeats() -
Method in class de.jstacs.classifier.assessment.RepeatedHoldOutAssessParameterSet
- Returns the repeats defined by this
RepeatedHoldOutAssessParameterSet (repeats define how many
iterations (train and test classifiers) of that
RepeatedHoldOutExperiment this
RepeatedHoldOutAssessParameterSet is used with are performed).
- getRepeats() -
Method in class de.jstacs.classifier.assessment.RepeatedSubSamplingAssessParameterSet
- Returns the repeats defined by this
RepeatedSubSamplingAssessParameterSet (repeats defines how many
iterations (train and test classifiers) of that
RepeatedSubSamplingExperiment this
RepeatedSubSamplingAssessParameterSet is used with are
performed).
- getRepeats() -
Method in class de.jstacs.classifier.assessment.Sampled_RepeatedHoldOutAssessParameterSet
- Returns the repeats defined by this
Sampled_RepeatedHoldOutAssessParameterSet (repeats defines how
many iterations (train and test classifiers) of that
Sampled_RepeatedHoldOutExperiment this
Sampled_RepeatedHoldOutAssessParameterSet is used with are
performed).
- getResult() -
Method in class de.jstacs.classifier.AbstractScoreBasedClassifier.DoubleTableResult
-
- getResult() -
Method in class de.jstacs.results.ImageResult
-
- getResult() -
Method in class de.jstacs.results.ListResult
-
- getResult() -
Method in class de.jstacs.results.Result
- Returns the value of the
Result.
- getResult() -
Method in class de.jstacs.results.SampleResult
-
- getResult() -
Method in class de.jstacs.results.SimpleResult
-
- getResult() -
Method in class de.jstacs.results.StorableResult
-
- getResult() -
Method in class de.jstacs.utils.galaxy.GalaxyAdaptor.FileResult
-
- getResultAt(int) -
Method in class de.jstacs.results.NumericalResultSet
-
- getResultAt(int) -
Method in class de.jstacs.results.ResultSet
- Returns
Result number index in this
ResultSet.
- getResultInstance() -
Method in class de.jstacs.results.StorableResult
- Returns the instance of the
Storable that is the result of this
StorableResult.
- getResults(Sample[], MeasureParameters, boolean, boolean) -
Method in class de.jstacs.classifier.AbstractClassifier
- This method computes the results for any evaluation of the classifier.
- getResults(Sample[], MeasureParameters, boolean, boolean) -
Method in class de.jstacs.classifier.AbstractScoreBasedClassifier
-
- getResults(Sample[], MeasureParameters, boolean, boolean) -
Method in class de.jstacs.classifier.MappingClassifier
-
- getResults() -
Method in class de.jstacs.results.ResultSet
- Returns all internal results as an array of
Results.
- getReverseComplementarySample() -
Method in class de.jstacs.data.Sample
- Returns a
Sample that contains the reverse complement of all Sequences in
this Sample.
- getReverseComplementDistributions(ComplementableDiscreteAlphabet, double[][][]) -
Static method in class de.jstacs.scoringFunctions.mix.StrandScoringFunction
- This method computes the reverse complement distributions for given conditional distributions.
- getReverseSwitches() -
Method in class de.jstacs.scoringFunctions.IndependentProductScoringFunction
- This method returns a deep copy of the internally used switches for the parts whether to use the corresponding
NormalizableScoringFunction forward or as reverse complement.
- getRNotation(String) -
Method in class de.jstacs.scoringFunctions.mix.motifSearch.DurationScoringFunction
- This method returns the distribution in R notation.
- getRNotation(String) -
Method in class de.jstacs.scoringFunctions.mix.motifSearch.MixtureDuration
-
- getRNotation(String) -
Method in class de.jstacs.scoringFunctions.mix.motifSearch.SkewNormalLikeScoringFunction
-
- getRNotation(String) -
Method in class de.jstacs.scoringFunctions.mix.motifSearch.UniformDurationScoringFunction
-
- getRoot() -
Method in class de.jstacs.models.phylo.PhyloTree
- This method returns the root node of the tree
- getRootValue(int) -
Method in class de.jstacs.algorithms.graphs.tensor.AsymmetricTensor
-
- getRootValue(int) -
Method in class de.jstacs.algorithms.graphs.tensor.SubTensor
-
- getRootValue(int) -
Method in class de.jstacs.algorithms.graphs.tensor.SymmetricTensor
-
- getRootValue(int) -
Method in class de.jstacs.algorithms.graphs.tensor.Tensor
- Returns the value for
child as root.
- getRunTimeException(Exception) -
Static method in class de.jstacs.models.hmm.AbstractHMM
- This method creates an
RuntimeException from any other Exception
- getSafeOutputStream(OutputStream) -
Static method in class de.jstacs.utils.SafeOutputStream
- This method returns an instance of
SafeOutputStream for a given OutputStream.
- getSample() -
Method in class de.jstacs.data.Sample.WeightedSampleFactory
- Returns the
Sample, where each Sequence occurs only
once.
- getSample(AlphabetContainer, AbstractStringExtractor...) -
Static method in class de.jstacs.data.sequences.SparseSequence
- This method allows to create a
Sample containing SparseSequences.
- getSampleForProperty(Sample, DinucleotideProperty) -
Static method in enum de.jstacs.data.DinucleotideProperty
- Creates a new
Sample by converting each Sequence in original to the DinucleotideProperty property.
- getSampleForProperty(Sample, DinucleotideProperty.Smoothing, boolean, DinucleotideProperty) -
Static method in enum de.jstacs.data.DinucleotideProperty
- Creates a new
Sample by converting each Sequence in original to the DinucleotideProperty property using the DinucleotideProperty.Smoothing smoothing.
- getSampleForProperty(Sample, DinucleotideProperty...) -
Static method in enum de.jstacs.data.DinucleotideProperty
- Creates a new
Sample by converting each Sequence in original to the DinucleotidePropertys properties and setting these as ReferenceSequenceAnnotation of each original sequence.
- getSampleForProperty(Sample, DinucleotideProperty.Smoothing, boolean, DinucleotideProperty...) -
Static method in enum de.jstacs.data.DinucleotideProperty
- Creates a new
Sample by converting each Sequence in original to the DinucleotidePropertys properties and adding or setting these as ReferenceSequenceAnnotation of each original sequence.
- getSamplingComponent() -
Method in class de.jstacs.classifier.scoringFunctionBased.sampling.SamplingScoreBasedClassifier
- Returns a sampling component suited for this
SamplingScoreBasedClassifier
- getSamplingGroups(int) -
Method in class de.jstacs.models.hmm.models.DifferentiableHigherOrderHMM
-
- getSamplingGroups(int) -
Method in class de.jstacs.scoringFunctions.directedGraphicalModels.MutableMarkovModelScoringFunction
-
- getSamplingGroups(int) -
Method in interface de.jstacs.scoringFunctions.SamplingScoringFunction
- Returns groups of indexes of parameters that shall be drawn
together in a sampling procedure
- getSamplingScheme() -
Method in class de.jstacs.classifier.scoringFunctionBased.sampling.SamplingScoreBasedClassifierParameterSet
- Returns the sampling scheme
- getScale() -
Method in class de.jstacs.parameters.RangeParameter
- Returns a description of the the scale of a range of parameter values.
- getScatterplot(AbstractScoreBasedClassifier, AbstractScoreBasedClassifier, Sample, Sample, REnvironment, boolean) -
Static method in class de.jstacs.classifier.utils.ClassificationVisualizer
- This method returns an
ImageResult containing a scatter plot of
the scores for the given classifiers cl1 and
cl2.
- getScore(Tensor, int[][]) -
Static method in class de.jstacs.algorithms.graphs.DAG
- Returns the score for any graph.
- getScore(Sequence, int) -
Method in class de.jstacs.classifier.AbstractScoreBasedClassifier
- This method returns the score for a given
Sequence and a given
class.
- getScore(Sequence, int, boolean) -
Method in class de.jstacs.classifier.AbstractScoreBasedClassifier
- This method returns the score for a given
Sequence and a given
class.
- getScore(Sequence, int, boolean) -
Method in class de.jstacs.classifier.MappingClassifier
-
- getScore(Sequence, int, boolean) -
Method in class de.jstacs.classifier.modelBased.ModelBasedClassifier
-
- getScore(Sequence, int, boolean) -
Method in class de.jstacs.classifier.scoringFunctionBased.sampling.SamplingScoreBasedClassifier
-
- getScore(Sequence, int, boolean) -
Method in class de.jstacs.classifier.scoringFunctionBased.ScoreClassifier
-
- getScoreForBestRun() -
Method in class de.jstacs.models.mixture.AbstractMixtureModel
- Returns the value of the optimized function from the best run of the last
training.
- getScoreForPath(Tensor, int, byte, int[]) -
Static method in class de.jstacs.algorithms.graphs.DAG
- Returns the score for a given path
path using the first
l nodes and dependencies of order k.
- getScores(Sample) -
Method in class de.jstacs.classifier.AbstractScoreBasedClassifier
- This method returns the scores of the classifier for any
Sequence
in the Sample.
- getScores(Sample) -
Method in class de.jstacs.classifier.modelBased.ModelBasedClassifier
-
- getScores(Sample) -
Method in class de.jstacs.classifier.scoringFunctionBased.sampling.SamplingScoreBasedClassifier
-
- getScoringFunction(int) -
Method in class de.jstacs.classifier.scoringFunctionBased.ScoreClassifier
- Returns the internally used
ScoringFunction with index
i.
- getScoringFunctions() -
Method in class de.jstacs.classifier.scoringFunctionBased.ScoreClassifier
- Returns all internally used
ScoringFunctions in the internal
order.
- getScoringFunctions() -
Method in class de.jstacs.scoringFunctions.mix.AbstractMixtureScoringFunction
- Returns a deep copy of all internal used
ScoringFunctions.
- getSecondElement() -
Method in class de.jstacs.utils.Pair
- This method returns the second element.
- getSelected() -
Method in class de.jstacs.parameters.CollectionParameter
- Returns the index of the selected value.
- getSelected() -
Method in class de.jstacs.parameters.MultiSelectionCollectionParameter
-
- getSensitivityForSpecificity(double[], double[], double) -
Static method in class de.jstacs.classifier.ScoreBasedPerformanceMeasureDefinitions
- This method computes the sensitivity for a given specificity.
- getSequenceAnnotationByType(String, int) -
Method in class de.jstacs.data.Sequence
- Returns the
SequenceAnnotation no.
- getSequenceAnnotationByTypeAndIdentifier(String, String) -
Method in class de.jstacs.data.Sequence
- Returns the
SequenceAnnotation of this Sequence that has type type and identifier identifier.
- getSequenceAnnotationIndexMatrix(String, Hashtable<String, Integer>, String, Hashtable<String, Integer>) -
Method in class de.jstacs.data.Sample
- This method creates a matrix which contains the index of the
Sequence with specific SequenceAnnotation
combination or -1 if the Sample does not contain any Sequence with such a combination.
- getSequenceWeights() -
Method in class de.jstacs.classifier.scoringFunctionBased.AbstractOptimizableFunction
-
- getSequenceWeights() -
Method in class de.jstacs.classifier.scoringFunctionBased.OptimizableFunction
- Returns the weights for each
Sequence for each
class used in this OptimizableFunction.
- getShannonEntropy(Model, int) -
Static method in class de.jstacs.models.utils.ModelTester
- This method computes the Shannon entropy for any discrete model
m and all sequences of length, if possible.
- getShannonEntropyInBits(Model, int) -
Static method in class de.jstacs.models.utils.ModelTester
- This method computes the Shannon entropy in bits for any discrete model
m and all sequences of length, if possible.
- getShortFromParameter(Parameter) -
Static method in class de.jstacs.io.ParameterSetParser
- Returns the
short which is the value of the
Parameter par.
- getSimplifiedAlphabetContainer(Alphabet[], int[]) -
Static method in class de.jstacs.data.AlphabetContainer
- This method creates a new
AlphabetContainer that uses as less as
possible Alphabets to describe the container.
- getSizeOfEventSpace() -
Method in class de.jstacs.models.hmm.states.emissions.continuous.GaussianEmission
-
- getSizeOfEventSpace() -
Method in interface de.jstacs.models.hmm.states.emissions.DifferentiableEmission
- Returns the size of the event space, i.e., the number of possible outcomes,
for the random variables of this emission
- getSizeOfEventSpace() -
Method in class de.jstacs.models.hmm.states.emissions.discrete.AbstractConditionalDiscreteEmission
-
- getSizeOfEventSpace() -
Method in class de.jstacs.models.hmm.states.emissions.SilentEmission
-
- getSizeOfEventSpace(int) -
Method in interface de.jstacs.models.hmm.transitions.DifferentiableTransition
- Returns the size of the event space, i.e., the number of possible outcomes,
for the random variable of parameter
index
- getSizeOfEventSpace(int) -
Method in class de.jstacs.models.hmm.transitions.HigherOrderTransition
-
- getSizeOfEventSpaceForRandomVariablesOfParameter(int) -
Method in class de.jstacs.models.hmm.models.DifferentiableHigherOrderHMM
-
- getSizeOfEventSpaceForRandomVariablesOfParameter(int) -
Method in class de.jstacs.scoringFunctions.CMMScoringFunction
-
- getSizeOfEventSpaceForRandomVariablesOfParameter(int) -
Method in class de.jstacs.scoringFunctions.directedGraphicalModels.BayesianNetworkScoringFunction
-
- getSizeOfEventSpaceForRandomVariablesOfParameter(int) -
Method in class de.jstacs.scoringFunctions.homogeneous.HMM0ScoringFunction
-
- getSizeOfEventSpaceForRandomVariablesOfParameter(int) -
Method in class de.jstacs.scoringFunctions.homogeneous.HMMScoringFunction
-
- getSizeOfEventSpaceForRandomVariablesOfParameter(int) -
Method in class de.jstacs.scoringFunctions.homogeneous.UniformHomogeneousScoringFunction
-
- getSizeOfEventSpaceForRandomVariablesOfParameter(int) -
Method in class de.jstacs.scoringFunctions.IndependentProductScoringFunction
-
- getSizeOfEventSpaceForRandomVariablesOfParameter(int) -
Method in class de.jstacs.scoringFunctions.MappingScoringFunction
-
- getSizeOfEventSpaceForRandomVariablesOfParameter(int) -
Method in class de.jstacs.scoringFunctions.mix.AbstractMixtureScoringFunction
-
- getSizeOfEventSpaceForRandomVariablesOfParameter(int) -
Method in class de.jstacs.scoringFunctions.mix.motifSearch.DurationScoringFunction
-
- getSizeOfEventSpaceForRandomVariablesOfParameter(int) -
Method in class de.jstacs.scoringFunctions.MRFScoringFunction
-
- getSizeOfEventSpaceForRandomVariablesOfParameter(int) -
Method in interface de.jstacs.scoringFunctions.NormalizableScoringFunction
- Returns the size of the event space of the random variables that are
affected by parameter no.
- getSizeOfEventSpaceForRandomVariablesOfParameter(int) -
Method in class de.jstacs.scoringFunctions.NormalizedScoringFunction
-
- getSizeOfEventSpaceForRandomVariablesOfParameter(int) -
Method in class de.jstacs.scoringFunctions.UniformScoringFunction
-
- getSmoothedProfile(int, String...) -
Method in class de.jstacs.motifDiscovery.KMereStatistic
- This method returns an array of smoothed profiles.
- getSmoothedProfile(int, Sequence...) -
Method in class de.jstacs.motifDiscovery.KMereStatistic
- This method returns an array of smoothed profiles.
- getSortedInitialParameters(ScoringFunction[], MutableMotifDiscovererToolbox.InitMethodForScoringFunction[], SFBasedOptimizableFunction, int, OutputStream, int) -
Static method in class de.jstacs.motifDiscovery.MutableMotifDiscovererToolbox
- This method allows to initialize the
MutableMotifDiscovererToolbox.InitMethodForScoringFunction using different MutableMotifDiscovererToolbox.InitMethodForScoringFunction.
- getSortedScoresForMotifAndFlanking(Sample, Sample, String) -
Static method in class de.jstacs.motifDiscovery.MotifDiscoveryAssessment
- Returns the scores read from the prediction
pred for the motif with identifier identifier and flanking sequences as annotated in
the Sample data.
- getSortedValuesForMotifAndFlanking(Sample, double[][], double, double, String) -
Static method in class de.jstacs.motifDiscovery.MotifDiscoveryAssessment
- This method provides some score arrays that can be used in
ScoreBasedPerformanceMeasureDefinitions to determine some
curves or area under curves based on the values of the predictions.
- getStartDistance() -
Method in class de.jstacs.models.hmm.training.NumericalHMMTrainingParameterSet
- This method returns the start distance that should be used in the line search during the optimization.
- getStartIndexOfAlignmentForFirst() -
Method in class de.jstacs.algorithms.alignment.PairwiseStringAlignment
- This method returns the start index of the alignment in the first sequence.
- getStartNode() -
Method in class de.jstacs.algorithms.graphs.Edge
- Returns the start node of the edge.
- getStartPositions(int, Sample, int, int) -
Method in class de.jstacs.motifDiscovery.SignificantMotifOccurrencesFinder
- This method returns a list of start positions of binding sites.
- getStartValue() -
Method in class de.jstacs.parameters.RangeParameter
- Returns the start value of a range of parameter values or
null if no range was specified.
- getStatePosteriorMatrixFor(Sequence) -
Method in class de.jstacs.models.hmm.AbstractHMM
- This method returns the log state posterior of all states for a sequence.
- getStatePosteriorMatrixFor(Sample) -
Method in class de.jstacs.models.hmm.AbstractHMM
- This method returns the state posteriors for all sequences of the sample
data.
- getStationaryDistribution(double[], int) -
Static method in class de.jstacs.models.utils.StationaryDistribution
- This method return the stationary distribution.
- getStatistics() -
Method in class de.jstacs.results.MeanResultSet
- Returns the means and (if possible the) standard errors of the results in
this
MeanResultSet as a new NumericalResultSet.
- getStatistics(Sample, double[], int, double) -
Static method in class de.jstacs.scoringFunctions.directedGraphicalModels.structureLearning.measures.Measure
- Counts the occurrences of symbols of the
AlphabetContainer of
Sample s using weights.
- getStatisticsOrderTwo(Sample, double[], int, double) -
Static method in class de.jstacs.scoringFunctions.directedGraphicalModels.structureLearning.measures.Measure
- Counts the occurrences of symbols of the
AlphabetContainer of
Sample s using weights.
- getSteps() -
Method in class de.jstacs.parameters.RangeParameter
- Returns the number of steps of a range of parameter values or
0 if no range was specified.
- getStrand(Sequence, int) -
Method in class de.jstacs.scoringFunctions.mix.StrandScoringFunction
- This method returns the preferred
StrandedLocatedSequenceAnnotationWithLength.Strand for a given subsequence.
- getStrand(Sequence, int) -
Method in class de.jstacs.scoringFunctions.NormalizedScoringFunction
- This method return the preferred
StrandedLocatedSequenceAnnotationWithLength.Strand for a Sequence beginning at startPos.
- getStrandedness() -
Method in class de.jstacs.data.sequences.annotation.StrandedLocatedSequenceAnnotationWithLength
- Returns the strandedness, i.e the orientation of this annotation.
- getStrandProbabilitiesFor(int, int, Sequence, int) -
Method in class de.jstacs.models.mixture.motif.SingleHiddenMotifMixture
-
- getStrandProbabilitiesFor(int, int, Sequence, int) -
Method in interface de.jstacs.motifDiscovery.MotifDiscoverer
- This method returns the probabilities of the strand orientations for a given subsequence if it is
considered as site of the motif model in a specific component.
- getStrandProbabilitiesFor(int, int, Sequence, int) -
Method in class de.jstacs.scoringFunctions.IndependentProductScoringFunction
-
- getStrandProbabilitiesFor(int, int, Sequence, int) -
Method in class de.jstacs.scoringFunctions.MappingScoringFunction
-
- getStrandProbabilitiesFor(int, int, Sequence, int) -
Method in class de.jstacs.scoringFunctions.mix.MixtureScoringFunction
-
- getStrandProbabilitiesFor(int, int, Sequence, int) -
Method in class de.jstacs.scoringFunctions.mix.motifSearch.HiddenMotifsMixture
-
- getStringFromParameter(Parameter) -
Static method in class de.jstacs.io.ParameterSetParser
- Returns the
String which is the value of the Parameter
par.
- getStringRepresentation(Object) -
Method in class de.jstacs.data.Sequence
- This method creates a String representation from the given representation.
- getStringRepresentation(Object) -
Method in class de.jstacs.data.Sequence.RecursiveSequence
-
- getStringRepresentation(Object) -
Method in class de.jstacs.data.sequences.ArbitrarySequence
-
- getStringRepresentation(Object) -
Method in class de.jstacs.data.sequences.MultiDimensionalDiscreteSequence
-
- getStringRepresentation(Object) -
Method in class de.jstacs.data.sequences.SimpleDiscreteSequence
-
- getStructure() -
Method in class de.jstacs.models.discrete.inhomogeneous.DAGModel
-
- getStructure() -
Method in class de.jstacs.models.discrete.inhomogeneous.FSDAGModel
-
- getStructure() -
Method in class de.jstacs.models.discrete.inhomogeneous.InhomogeneousDGM
- Returns a
String representation of the underlying graph.
- getStructure() -
Method in class de.jstacs.models.discrete.inhomogeneous.shared.SharedStructureMixture
- Returns a
String representation of the structure of the used
models.
- getStructure(Sample, double[], StructureLearner.ModelType, byte, StructureLearner.LearningType) -
Method in class de.jstacs.models.discrete.inhomogeneous.StructureLearner
- This method finds the optimal structure of a model by using a given
learning method (in some sense).
- getStructure(Tensor, StructureLearner.ModelType, byte) -
Static method in class de.jstacs.models.discrete.inhomogeneous.StructureLearner
- This method can be used to determine the optimal structure of a model.
- getStructureFromPath(int[], Tensor) -
Static method in class de.jstacs.algorithms.graphs.DAG
- Extracts the structure from a given path
path and
score-"function".
- getSubAnnotations() -
Method in class de.jstacs.data.sequences.annotation.SequenceAnnotation
- Returns the sub-annotations of this
SequenceAnnotation as given
in the constructor.
- getSubContainer(int, int) -
Method in class de.jstacs.data.AlphabetContainer
- Returns a sub-container with the
Alphabets for the positions
starting at start and with length length.
- getSubSequence(AlphabetContainer, int) -
Method in class de.jstacs.data.Sequence
- This method should be used if one wants to create a
Sample of
subsequences of defined length.
- getSubSequence(AlphabetContainer, int, int) -
Method in class de.jstacs.data.Sequence
- This method should be used if one wants to create a
Sample of
subsequences of defined length.
- getSubSequence(int) -
Method in class de.jstacs.data.Sequence
- This is a very efficient way to create a subsequence/suffix for
Sequences with a simple AlphabetContainer.
- getSubSequence(int, int) -
Method in class de.jstacs.data.Sequence
- This is a very efficient way to create a subsequence of defined length
for
Sequences with a simple AlphabetContainer.
- getSuffixSample(int) -
Method in class de.jstacs.data.Sample
- This method enables you to use only a suffix of all elements, i.e. the
Sequence, in the current Sample.
- getSumOfDeviation(Model, Model, int) -
Static method in class de.jstacs.models.utils.ModelTester
- This method computes the sum of deviations between the probabilities for
all sequences of
length for discrete models m1
and m2.
- getSumOfDistribution(Model, int) -
Static method in class de.jstacs.models.utils.ModelTester
- This method computes the marginal distribution for any discrete model
m and all sequences of length, if possible.
- getSumOfHyperparameter() -
Method in class de.jstacs.utils.random.DirichletMRGParams
- Returns the sum of the hyperparameters (entries of the hyperparameter
vector) of the underlying Dirichlet distribution.
- getSumOfHyperparameter() -
Method in class de.jstacs.utils.random.ErlangMRGParams
- Returns the sum of the hyperparameters (entries of the hyperparameter
vector) of the underlying Erlang distribution.
- getSumOfHyperParameters(int, int, double) -
Static method in class de.jstacs.scoringFunctions.homogeneous.HMMScoringFunction
- This method returns an array that can be used in the constructor
HMMScoringFunction.HMMScoringFunction(AlphabetContainer, int, double, double[], boolean, boolean, int)
containing the sums of the specific hyperparameters.
- getSumOfWeights() -
Method in class de.jstacs.data.Sample.WeightedSampleFactory
- Returns the sum of all weights.
- getSuperClassOf(T...) -
Static method in class de.jstacs.io.ArrayHandler
- This method returns the deepest class in the class hierarchy that is the
class or a superclass of all instances in the array.
- getSymbol(int, double) -
Method in class de.jstacs.data.AlphabetContainer
- Returns a
String representation of the encoded symbol
val of the Alphabet of position pos of
this AlphabetContainer.
- getSymbolAt(int) -
Method in class de.jstacs.data.alphabets.DiscreteAlphabet
- Returns the symbol at position
i in the alphabet.
- getSymKLDivergence(Model, Model, int) -
Static method in class de.jstacs.models.utils.ModelTester
- Returns the difference of the Kullback-Leibler-divergences, i.e.
- getTempDir() -
Method in class de.jstacs.classifier.scoringFunctionBased.sampling.SamplingScoreBasedClassifier
- Returns the directory for parameter files set in this
SamplingScoreBasedClassifier.
- getTensor(Sample, double[], byte, StructureLearner.LearningType) -
Method in class de.jstacs.models.discrete.inhomogeneous.StructureLearner
- This method can be used to compute a
Tensor that can be used to
determine the optimal structure.
- getTerminantionCondition() -
Method in class de.jstacs.models.hmm.training.MaxHMMTrainingParameterSet
- This method returns the
AbstractTerminationCondition for stopping the training, e.g., if the
difference of the scores between two iterations is smaller than a given
threshold the training is stopped.
- getThreshold() -
Method in class de.jstacs.classifier.ScoreBasedPerformanceMeasureDefinitions.ThresholdMeasurePair
- This method returns the value of threshold.
- getThreshold(double[], int) -
Static method in class de.jstacs.classifier.utils.PValueComputation
- This method returns the threshold that determines if an observed score is
significant.
- getThreshold() -
Method in class de.jstacs.sampling.VarianceRatioBurnInTestParameterSet
- Returns the threshold used in the
VarianceRatioBurnInTestParameterSet.
- getTime() -
Method in class de.jstacs.models.mixture.AbstractMixtureModel
- This method returns an instance of
Time that is used for the TerminationCondition in the EM.
- getTopologicalOrder(int[][]) -
Static method in class de.jstacs.algorithms.graphs.TopSort
- Returns the topological order of indexes according to
parents2.
- getTopologicalOrder2(byte[][]) -
Static method in class de.jstacs.algorithms.graphs.TopSort
- Computes a topological ordering for a given graph.
- getTrain_TestNumbers(boolean) -
Method in class de.jstacs.classifier.assessment.RepeatedSubSamplingAssessParameterSet
- Returns an array containing the number of elements the subsampled (train
| test) datasets should consist of.
- getTransitionElementIndex(int, int) -
Method in class de.jstacs.models.hmm.transitions.BasicHigherOrderTransition
- This method return the index of the
BasicHigherOrderTransition.AbstractTransitionElement using the BasicHigherOrderTransition.lookup table.
- getTrueIndexForLastGetBest() -
Method in class de.jstacs.algorithms.graphs.tensor.SymmetricTensor
- Returns the edge from
SymmetricTensor.getBest(int, int[], byte) in an encoded
index.
- getType() -
Method in class de.jstacs.data.AlphabetContainer
- Returns the type of this
AlphabetContainer.
- getType() -
Method in enum de.jstacs.data.DinucleotideProperty
- Returns the type of this property.
- getType() -
Method in class de.jstacs.data.sequences.annotation.SequenceAnnotation
- Returns the type of this
SequenceAnnotation as given in the
constructor.
- getValidator() -
Method in class de.jstacs.parameters.SimpleParameter
- Returns the
ParameterValidator used in this
SimpleParameter.
- getValue(byte, int, int...) -
Method in class de.jstacs.algorithms.graphs.tensor.AsymmetricTensor
-
- getValue(byte, int, int...) -
Method in class de.jstacs.algorithms.graphs.tensor.SubTensor
-
- getValue(byte, int, int...) -
Method in class de.jstacs.algorithms.graphs.tensor.SymmetricTensor
-
- getValue(byte, int, int...) -
Method in class de.jstacs.algorithms.graphs.tensor.Tensor
- Returns the value for the edge
parents[0],...
- getValue() -
Method in class de.jstacs.parameters.CollectionParameter
-
- getValue() -
Method in class de.jstacs.parameters.EnumParameter
-
- getValue() -
Method in class de.jstacs.parameters.FileParameter
-
- getValue() -
Method in class de.jstacs.parameters.MultiSelectionCollectionParameter
-
- getValue() -
Method in class de.jstacs.parameters.Parameter
- Returns the current value of this
Parameter.
- getValue() -
Method in class de.jstacs.parameters.ParameterSetContainer
-
- getValue() -
Method in class de.jstacs.parameters.RangeParameter
-
- getValue() -
Method in class de.jstacs.parameters.SimpleParameter
-
- getValue() -
Method in class de.jstacs.scoringFunctions.directedGraphicalModels.Parameter
- Returns the current value of this parameter.
- getValueFor(String) -
Method in class de.jstacs.parameters.MultiSelectionCollectionParameter
- Returns the value for the option with key
key.
- getValueFor(int) -
Method in class de.jstacs.parameters.MultiSelectionCollectionParameter
- Returns the value of the option with no.
- getValueFromTag(String) -
Method in class de.jstacs.parameters.ParameterSetTagger
- This method returns the value of the
Parameter specified by the tag.
- getValueFromTag(String, Class<T>) -
Method in class de.jstacs.parameters.ParameterSetTagger
- This method returns the casted value of the
Parameter specified by the tag.
- getValueOfAIC(Model, Sample, int) -
Static method in class de.jstacs.models.utils.ModelTester
- This method computes the value of Akaikes Information Criterion (AIC).
- getValueOfBIC(Model, Sample, int) -
Static method in class de.jstacs.models.utils.ModelTester
- This method computes the value of the Bayesian Information Criterion
(BIC).
- getValues() -
Method in class de.jstacs.parameters.MultiSelectionCollectionParameter
- Returns the values of all selected options as an array.
- getValuesForEachNucleotide(Sample, int, boolean) -
Method in class de.jstacs.motifDiscovery.SignificantMotifOccurrencesFinder
- This method determines a score for each possible starting position in each of the sequences in
data
that this position is covered by at least one motif occurrence of the
motif with index index.
- getValuesFromSequence(Sequence, int) -
Method in class de.jstacs.scoringFunctions.mix.motifSearch.PositionScoringFunction
- This method extracts the values form a sequence.
- getVersionInformation() -
Method in class de.jstacs.utils.REnvironment
- Returns information about the version of R that is used.
- getViterbiPath(int, int, Sequence, SamplingHigherOrderHMM.ViterbiComputation) -
Method in class de.jstacs.models.hmm.models.SamplingHigherOrderHMM
- This method returns a viterbi path that is the optimum for the choosen ViterbiComputation method
- getViterbiPathFor(int, int, Sequence) -
Method in class de.jstacs.models.hmm.AbstractHMM
-
- getViterbiPathFor(Sequence) -
Method in class de.jstacs.models.hmm.AbstractHMM
-
- getViterbiPathFor(int, int, Sequence) -
Method in class de.jstacs.models.hmm.models.HigherOrderHMM
-
- getViterbiPathFor(int, int, Sequence) -
Method in class de.jstacs.models.hmm.models.SamplingHigherOrderHMM
-
- getViterbiPathsFor(Sample) -
Method in class de.jstacs.models.hmm.AbstractHMM
- This method returns the viterbi paths and scores for all sequences of the sample
data.
- getWeight() -
Method in class de.jstacs.algorithms.graphs.Edge
- Returns the weight of the edge.
- getWeight(int) -
Method in class de.jstacs.data.Sample.WeightedSampleFactory
- Returns the weight for the
Sequence with index
index.
- getWeight() -
Method in class de.jstacs.models.phylo.PhyloNode
- This method return the weight (length, rate ...) for the incoming edge
- getWeight() -
Method in class de.jstacs.utils.ComparableElement
- This method returns the weight of the element.
- getWeights() -
Method in class de.jstacs.data.Sample.WeightedSampleFactory
- Returns a copy of the weights for the
Sample.
- getWeights() -
Method in class de.jstacs.models.mixture.AbstractMixtureModel
- This method returns a deep copy of the weights for each component.
- getWriter() -
Method in class de.jstacs.utils.galaxy.GalaxyAdaptor.Protocol
- Returns the
PrintWriter of this protocol
- getXmlTag() -
Method in class de.jstacs.algorithms.optimization.termination.AbsoluteValueCondition
- Deprecated.
- getXmlTag() -
Method in class de.jstacs.algorithms.optimization.termination.AbstractTerminationCondition
- This method returns the xml tag that is used in the method
AbstractTerminationCondition.toXML() and
in the constructor AbstractTerminationCondition.AbstractTerminationCondition(StringBuffer).
- getXmlTag() -
Method in class de.jstacs.algorithms.optimization.termination.CombinedCondition
-
- getXmlTag() -
Method in class de.jstacs.algorithms.optimization.termination.IterationCondition
-
- getXmlTag() -
Method in class de.jstacs.algorithms.optimization.termination.SmallDifferenceOfFunctionEvaluationsCondition
-
- getXmlTag() -
Method in class de.jstacs.algorithms.optimization.termination.SmallGradientConditon
-
- getXmlTag() -
Method in class de.jstacs.algorithms.optimization.termination.SmallStepCondition
-
- getXmlTag() -
Method in class de.jstacs.algorithms.optimization.termination.TimeCondition
-
- getXMLTag() -
Method in class de.jstacs.classifier.AbstractClassifier
- Returns the
String that is used as tag for the XML representation
of the classifier.
- getXMLTag() -
Method in class de.jstacs.classifier.MappingClassifier
-
- getXMLTag() -
Method in class de.jstacs.classifier.modelBased.ModelBasedClassifier
-
- getXMLTag() -
Method in class de.jstacs.classifier.scoringFunctionBased.gendismix.GenDisMixClassifier
-
- getXMLTag() -
Method in class de.jstacs.classifier.scoringFunctionBased.msp.MSPClassifier
-
- getXMLTag() -
Method in class de.jstacs.classifier.scoringFunctionBased.sampling.SamplingGenDisMixClassifier
-
- getXMLTag() -
Method in class de.jstacs.classifier.scoringFunctionBased.ScoreClassifier
-
- getXMLTag() -
Method in class de.jstacs.models.discrete.Constraint
- Returns the XML tag that is used for the class to en- or decode.
- getXMLTag() -
Method in class de.jstacs.models.discrete.DiscreteGraphicalModel
- Returns the XML tag that is used for this model in
DiscreteGraphicalModel.fromXML(StringBuffer) and DiscreteGraphicalModel.toXML().
- getXMLTag() -
Method in class de.jstacs.models.discrete.homogeneous.HomogeneousMM
-
- getXMLTag() -
Method in class de.jstacs.models.discrete.homogeneous.HomogeneousModel.HomCondProb
-
- getXMLTag() -
Method in class de.jstacs.models.discrete.inhomogeneous.BayesianNetworkModel
-
- getXMLTag() -
Method in class de.jstacs.models.discrete.inhomogeneous.FSDAGModel
-
- getXMLTag() -
Method in class de.jstacs.models.discrete.inhomogeneous.InhCondProb
-
- getXMLTag() -
Method in class de.jstacs.models.discrete.inhomogeneous.MEMConstraint
-
- getXMLTag() -
Method in class de.jstacs.models.hmm.AbstractHMM
- Returns the tag for the XML representation.
- getXMLTag() -
Method in class de.jstacs.models.hmm.models.HigherOrderHMM
-
- getXMLTag() -
Method in class de.jstacs.models.hmm.models.SamplingHigherOrderHMM
-
- getXMLTag() -
Method in class de.jstacs.models.hmm.transitions.BasicHigherOrderTransition.AbstractTransitionElement
- This method returns the xml tag used in
BasicHigherOrderTransition.AbstractTransitionElement.toXML().
- getXMLTag() -
Method in class de.jstacs.models.hmm.transitions.BasicHigherOrderTransition
- The method returns the XML tag used during saving and loading the transition.
- getXMLTag() -
Method in class de.jstacs.models.hmm.transitions.elements.BasicPluginTransitionElement
-
- getXMLTag() -
Method in class de.jstacs.models.hmm.transitions.elements.DistanceBasedScaledTransitionElement
-
- getXMLTag() -
Method in class de.jstacs.models.hmm.transitions.elements.ScaledTransitionElement
-
- getXMLTag() -
Method in class de.jstacs.models.hmm.transitions.HigherOrderTransition
-
- getXMLTag() -
Method in class de.jstacs.sampling.AbstractBurnInTest
- This method returns the XML tag that is used in
AbstractBurnInTest.toXML() and
AbstractBurnInTest.AbstractBurnInTest(StringBuffer).
- getXMLTag() -
Method in class de.jstacs.sampling.VarianceRatioBurnInTest
-
- getXMLTag() -
Method in class de.jstacs.scoringFunctions.mix.AbstractMixtureScoringFunction
- This method returns the XML tag of the instance that is used to build a
XML representation.
- gibbsSampling(int, int, double, Sequence) -
Method in class de.jstacs.models.hmm.models.SamplingHigherOrderHMM
- This method implements a sampling step in the sampling procedure
- GibbsSamplingModel - Interface in de.jstacs.sampling
- This is the interface that any
AbstractModel has to implement if it
should be used in a sampling. - gibbsSamplingStep(int, int, boolean, Sample, double[]) -
Method in class de.jstacs.models.hmm.models.SamplingHigherOrderHMM
- This method implements the next step(s) in the sampling procedure
- goldenRatio(OneDimensionalFunction, double, double, double, double, double) -
Static method in class de.jstacs.algorithms.optimization.Optimizer
- Approximates a minimum (not necessary the global) in the interval
[lower,upper].
- goldenRatio(OneDimensionalFunction, double, double, double) -
Static method in class de.jstacs.algorithms.optimization.Optimizer
- Approximates a minimum (not necessary the global) in the interval
[lower,upper].
- grad -
Variable in class de.jstacs.models.hmm.states.emissions.discrete.AbstractConditionalDiscreteEmission
- The array for storing the gradients for
each parameter
- gradient -
Variable in class de.jstacs.models.hmm.models.DifferentiableHigherOrderHMM
- Help array for the gradient
- GT -
Static variable in interface de.jstacs.parameters.validation.Constraint
- The condition is greater than
- GUIProgressUpdater - Class in de.jstacs.utils
- This class implements a
ProgressUpdater with a GUI. - GUIProgressUpdater(boolean) -
Constructor for class de.jstacs.utils.GUIProgressUpdater
- This is the constructor for a
GUIProgressUpdater.