DAGModel).Storable.
enum defines a number of data types that can be used for
Parameters and Result
s.FileFilter that accepts Files that were modified after the date that is given in the constructor.Files that were modified after the given year, month, ... .
Files that were modified after d.
Classifiers that are based on ModelsClassifiers that are based on ScoringFunctions.NormalizableScoringFunctions by
a unified generative-discriminative learning principleScoringFunctions either
by maximum supervised posterior (MSP) or by maximum conditional likelihood (MCL)AbstractScoreBasedClassifiers that are based on SamplingScoringFunctions and that sample parameters
using the Metropolis-Hastings algorithm.DNAAlphabet for the most common case of DNA-sequencesSequences using a number of pre-defined annotation types, or additional
implementations of the SequenceAnnotation classStorables to an XML-representationModel and its abstract implementation AbstractModel, which is the super class of all other models.AbstractHMMParameter-interface.Parameter valuesScoringFunctions that can be used in a ScoreClassifier.ScoringFunctions that are equivalent to directed graphical models.BayesianNetworkScoringFunction.BayesianNetworkScoringFunction as
a Bayesian tree using a number of measures to define a rating of structuresBayesianNetworkScoringFunction as
a permuted Markov model using a number of measures to define a rating of structuresScoringFunctions that are homogeneous, i.e. model probabilities or scores independent of the position within a sequenceScoringFunctions that are mixtures of other ScoringFunctions.OutputStream.
ProgressUpdater and prints the
percentage of iterations that is already done on the screen.DefaultProgressUpdater.
MeasureParameters.setSelected(Measure, boolean)
for all MeasureParameters.Measures.
AbstractMixtureScoringFunction.isNormalized().
ParameterSet for any parameter set of
a DiscreteGraphicalModel.Storable.
length.
Sample data and
the Samples samples.
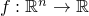 .
.HigherOrderHMM and a NormalizableScoringFunction by implementing some of the declared methods.Storable.
Optimizer.Exceptions depending on wrong dimensions of vectors
for a given function.DimensionException with standard error message
("The vector has wrong dimension for this function.
DimensionException with a more detailed error
message.
enum defines physicochemical, conformational, and letter-based dinucleotide properties of nucleotide sequences.DinucleotideProperty.MeanSmoothing that averages over windows of width width.
DinucleotideProperty.MedianSmoothing that computes the median over windows of width width.
DinucleotideProperty.Smoothing that conducts no smoothing.Strings.Storable.
InstantiableFromParameterSet
interface.
DiscreteAlphabet from a minimal and a maximal
value, i.e. in [min,max].
DiscreteAlphabet from a given alphabet as a
String array.
ParameterSet of a
DiscreteAlphabet.DiscreteAlphabet.
DiscreteAlphabet.DiscreteAlphabetParameterSet with empty values.
DiscreteAlphabet.DiscreteAlphabetParameterSet from an alphabet
given as a String array.
DiscreteAlphabet.DiscreteAlphabetParameterSet from an alphabet
of symbols given as a char array.
Storable
.
DiscreteAlphabet.DiscreteAlphabetMapping.
Storable.
DiscreteEmission based on the equivalent sample size.
DiscreteEmission defining the individual hyper parameters.
DiscreteEmission from its XML representation.
Storable.
Samples for discrete inhomogeneous models by a naive implementation.DiscreteSequence with the
AlphabetContainer container and the annotation
annotation but without the content.
Sequences of a specific
AlphabetContainer and length.DiscreteSequenceEnumerator from a given
AlphabetContainer and a length.
pos of the
Sequence.
Parameter, which is defined not to be free.
DoubleLists are used during the parallel computation of the gradient.
DoubleLists that are used while
computing the partial derivation.
Storable.
InstantiableFromParameterSet
interface.
DNAAlphabet.DNAAlphabet.DNAAlphabetParameterSet.
Storable
.
Samples of DNA Sequences.fName.
fName.
fName using the given parser.
Sequence seq fulfills all
requirements defined in the Parameter.context.
LogPrior that does not penalize any parameter.MutableMotifDiscovererToolbox.InitMethodForScoringFunction is a MutableMotifDiscoverer.
train-method
double.DoubleList with
initial length 10.
DoubleList with
initial length size.
Storable.
DoubleSymbolException is thrown if a symbol occurred more than once
in an alphabet.DoubleSymbolException that takes the symbol
that occurs more than once in the error message.
constr.
contrast and
endIdx-startIdx distributions drawn from a Dirichlet density centered around contrast, i.e. the hyper-parameters
of the Dirichlet density are the probabilities of contrast weighted by samples.
contrast[i] each weighted by weights[i]
kls.length distributions drawn from a Dirichlet density centered around contrast, i.e. the hyper-parameters
of the Dirichlet density are the probabilities of contrast weighted by samples.
ess (equivalent sample size)
as hyperparameters.
Storable.